+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Selenomonas sp. Cascade (SsCascade) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / Genome engineering / RNA-guided DNA endonuclease / Non-coding RNA / HNH nuclease / IMMUNE SYSTEM | |||||||||
| Biological species |  Selenomonas sp. (bacteria) / synthetic construct (others) Selenomonas sp. (bacteria) / synthetic construct (others) | |||||||||
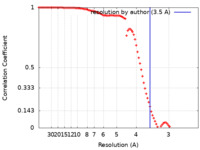
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Hirano S / Zhang F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Structural determinants of DNA cleavage by a CRISPR HNH-Cascade system. Authors: Seiichi Hirano / Han Altae-Tran / Soumya Kannan / Rhiannon K Macrae / Feng Zhang /  Abstract: Canonical prokaryotic type I CRISPR-Cas adaptive immune systems contain a multicomponent effector complex called Cascade, which degrades large stretches of DNA via Cas3 helicase-nuclease activity. ...Canonical prokaryotic type I CRISPR-Cas adaptive immune systems contain a multicomponent effector complex called Cascade, which degrades large stretches of DNA via Cas3 helicase-nuclease activity. Recently, a highly precise subtype I-F1 CRISPR-Cas system (HNH-Cascade) was found that lacks Cas3, the absence of which is compensated for by the insertion of an HNH endonuclease domain in the Cas8 Cascade component. Here, we describe the cryo-EM structure of Selenomonas sp. HNH-Cascade (SsCascade) in complex with target DNA and characterize its mechanism of action. The Cascade scaffold is complemented by the HNH domain, creating a ring-like structure in which the unwound target DNA is precisely cleaved. This structure visualizes a unique hybrid of two extensible biological systems-Cascade, an evolutionary platform for programmable DNA effectors, and an HNH nuclease, an adaptive domain with a spectrum of enzymatic activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43729.map.gz emd_43729.map.gz | 66.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43729-v30.xml emd-43729-v30.xml emd-43729.xml emd-43729.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43729_fsc.xml emd_43729_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_43729.png emd_43729.png | 73.6 KB | ||
| Filedesc metadata |  emd-43729.cif.gz emd-43729.cif.gz | 7.3 KB | ||
| Others |  emd_43729_half_map_1.map.gz emd_43729_half_map_1.map.gz emd_43729_half_map_2.map.gz emd_43729_half_map_2.map.gz | 65.1 MB 65.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43729 http://ftp.pdbj.org/pub/emdb/structures/EMD-43729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43729 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43729.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43729.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.202 Å | ||||||||||||||||||||||||||||||||||||
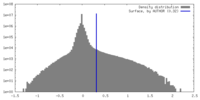
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43729_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
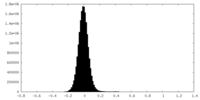
| Density Histograms |
-Half map: #1
| File | emd_43729_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
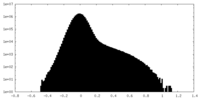
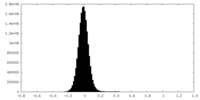
| Density Histograms |
- Sample components
Sample components
-Entire : Target DNA-bound SsCascade
| Entire | Name: Target DNA-bound SsCascade |
|---|---|
| Components |
|
-Supramolecule #1: Target DNA-bound SsCascade
| Supramolecule | Name: Target DNA-bound SsCascade / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
-Macromolecule #1: Cas7
| Macromolecule | Name: Cas7 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 38.700172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAANKKATNV TLKSRPENLS FARCLNTTEA KFWQTDFLKR HTFKLPLLIT DKAVLASKGH EMPPDKLEKE IMDPNPQKSQ SCTLSTECD TLRIDFGIKV LPVKESMYSC SDYNYRTAIY QKIDEYIAED GFLTLAKRYV NNIANARFLW RNRKGAEIIE T IVTIEDKE ...String: MAANKKATNV TLKSRPENLS FARCLNTTEA KFWQTDFLKR HTFKLPLLIT DKAVLASKGH EMPPDKLEKE IMDPNPQKSQ SCTLSTECD TLRIDFGIKV LPVKESMYSC SDYNYRTAIY QKIDEYIAED GFLTLAKRYV NNIANARFLW RNRKGAEIIE T IVTIEDKE YPSFNSKSFN LDTFVEDNAT INEIAQQIAD TFAGKREYLN IYVTCFVKIG CAMEVYPSQE MTFDDDDKGK KL FKFEGSA GMHSQKINNA LRTIDTWYPD YTTYEFPIPV ENYGAARSIG IPFRPDTKSF YKLIDRMILK NEDLPIEDKH YVM AILIRG GMFSKKQEK |
-Macromolecule #2: Cas8
| Macromolecule | Name: Cas8 / type: protein_or_peptide / ID: 2 Details: This protein is histidine tagged at N-terminal region from residues 1 to 41. Residues 42 to 385 are derived from natural Cas8 protein sequences. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 43.706289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRSKHHHHS GHHHTGHHHH SGSHHHTGGS GENLYFQGSG GLRNKILAAI SQKIPEEQKI NKYIEGLFQS IDKNHLATHV AKFTETNSP GNIGAYDILS SDMNCGYLDT ANAGWKEPDI VTNDAKYKRP QGFVAMEMSD GRTVMEHLQE DSAELRHEME E LTDKYDEI ...String: MLRSKHHHHS GHHHTGHHHH SGSHHHTGGS GENLYFQGSG GLRNKILAAI SQKIPEEQKI NKYIEGLFQS IDKNHLATHV AKFTETNSP GNIGAYDILS SDMNCGYLDT ANAGWKEPDI VTNDAKYKRP QGFVAMEMSD GRTVMEHLQE DSAELRHEME E LTDKYDEI RDGILNMPSM QPYRTNQFIK QVFFPVGGSY HLLSILPSTV LNYEVSDRLY RSKIPKIRLR LLSSNAASTT GS RLVSKNK WPLVFQALPP KFLEKNLAKA LDKEYLLPDI NIDELEGVDN GCLIDEALLP LIIDEGKRKG EGNYRPRHLR DER KEETVQ AFLDKYGYCN IPVGYEVHHI VPLSQGGADS IKNMIMLSIE HHERVTEAHA SYFKWRNT |
-Macromolecule #3: Cas5
| Macromolecule | Name: Cas5 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 28.703135 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMKGYILLEK VNIENANAFN NIIVGIPAIT SFLGFARALE RKLNAKEIAI RINGVGLEFH EYELKGYKNK RGQYVTSCPL PGSIPGQNE KKLDAHIMNQ AYIDLNMSFL LEVEGPHVDM STCKSIKSTM ETLRIAGGII RNYKKIRLID TLADIPYGYF L TLRQDNLN ...String: MMKGYILLEK VNIENANAFN NIIVGIPAIT SFLGFARALE RKLNAKEIAI RINGVGLEFH EYELKGYKNK RGQYVTSCPL PGSIPGQNE KKLDAHIMNQ AYIDLNMSFL LEVEGPHVDM STCKSIKSTM ETLRIAGGII RNYKKIRLID TLADIPYGYF L TLRQDNLN DAAGDDMLDK MIHALQQEDT LVPIAVGFKA LSEVGHVEGQ RDPEKDHCFV ESIFSLGGFE CSKILEDINS CL WRYKTEE GLYLCTII |
-Macromolecule #4: Cas6
| Macromolecule | Name: Cas6 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 20.735873 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFSQILIIKP GTGISPNIII SEDIFPVLHS LFVEHDKKFG ITFPAYSFDK KGHLGNIIEV LSEDKEALAS LCLEEHLAEV TDYVKVKKE ITFTDDYVLF KRIREENQYE TTARRMRKRG HTELGRPLEM HIKKKNQQIF CHAYIKVKSA STGQSYNIFL A PTDIKHGS FSAYGLLRGD THA |
-Macromolecule #5: Non-target strand DNA
| Macromolecule | Name: Non-target strand DNA / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 20.484176 KDa |
| Sequence | String: (DG)(DA)(DG)(DT)(DT)(DG)(DC)(DA)(DG)(DC) (DA)(DA)(DG)(DC)(DG)(DT)(DA)(DC)(DG)(DG) (DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA)(DT) (DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG)(DG)(DC) (DC) (DA)(DC)(DT)(DG)(DT)(DT) ...String: (DG)(DA)(DG)(DT)(DT)(DG)(DC)(DA)(DG)(DC) (DA)(DA)(DG)(DC)(DG)(DT)(DA)(DC)(DG)(DG) (DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA)(DT) (DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG)(DG)(DC) (DC) (DA)(DC)(DT)(DG)(DT)(DT)(DA)(DA) (DA)(DA)(DA)(DG)(DC)(DA)(DA)(DC)(DA)(DG) (DC)(DT) (DG)(DA)(DT)(DT)(DG)(DC) |
-Macromolecule #7: Target strand DNA
| Macromolecule | Name: Target strand DNA / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 20.20293 KDa |
| Sequence | String: (DG)(DC)(DA)(DA)(DT)(DC)(DA)(DG)(DC)(DT) (DG)(DT)(DT)(DG)(DC)(DT)(DT)(DT)(DT)(DT) (DA)(DA)(DC)(DA)(DG)(DT)(DG)(DG)(DC) (DC)(DT)(DT)(DA)(DT)(DT)(DA)(DA)(DA)(DT) (DG) (DA)(DC)(DT)(DT)(DC)(DT) ...String: (DG)(DC)(DA)(DA)(DT)(DC)(DA)(DG)(DC)(DT) (DG)(DT)(DT)(DG)(DC)(DT)(DT)(DT)(DT)(DT) (DA)(DA)(DC)(DA)(DG)(DT)(DG)(DG)(DC) (DC)(DT)(DT)(DA)(DT)(DT)(DA)(DA)(DA)(DT) (DG) (DA)(DC)(DT)(DT)(DC)(DT)(DC)(DC) (DG)(DT)(DA)(DC)(DG)(DC)(DT)(DT)(DG)(DC) (DT)(DG) (DC)(DA)(DA)(DC)(DT)(DC) |
-Macromolecule #6: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 19.746777 KDa |
| Sequence | String: UUUAGAAGGA GAAGUCAUUU AAUAAGGCCA CUGUUAAAAA GUGUACCGCC GGAUAGGCGG U |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.05 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)