+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of insect gustatory receptor BmGr9 | ||||||||||||
 Map data Map data | Map of unbound BmGr9 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | gustatory receptor / homotetramer / ligand-gated ion channel / seven transmembrane / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationionotropic taste receptor activity / detection of chemical stimulus involved in sensory perception of sweet taste / male courtship behavior / chemosensory behavior / ligand-gated monoatomic cation channel activity / monoatomic cation transmembrane transport / axon / neuronal cell body / dendrite / signal transduction / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
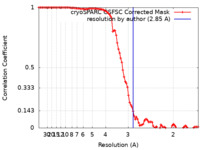
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | ||||||||||||
 Authors Authors | Frank HM / Walsh Jr RM / Garrity PA / Gaudet R | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Structure of an insect gustatory receptor. Authors: Heather M Frank / Sanket Walujkar / Richard M Walsh / Willem J Laursen / Douglas L Theobald / Paul A Garrity / Rachelle Gaudet /  Abstract: Gustatory Receptors (GRs) are critical for insect chemosensation and are potential targets for controlling pests and disease vectors. However, GR structures have not been experimentally determined. ...Gustatory Receptors (GRs) are critical for insect chemosensation and are potential targets for controlling pests and disease vectors. However, GR structures have not been experimentally determined. We present structures of Gr9 (BmGr9), a fructose-gated cation channel, in agonist-free and fructose-bound states. BmGr9 forms a tetramer similar to distantly related insect Olfactory Receptors (ORs). Upon fructose binding, BmGr9's ion channel gate opens through helix S7b movements. In contrast to ORs, BmGR9's ligand-binding pocket, shaped by a kinked helix S4 and a shorter extracellular S3-S4 loop, is larger and solvent accessible in both agonist-free and fructose-bound states. Also unlike ORs, fructose binding by BmGr9 involves helix S5 and a binding pocket lined with aromatic and polar residues. Structure-based sequence alignments reveal distinct patterns of ligand-binding pocket residue conservation in GR subfamilies associated with distinct ligand classes. These data provide insight into the molecular basis of GR ligand specificity and function. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43129.map.gz emd_43129.map.gz | 87.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43129-v30.xml emd-43129-v30.xml emd-43129.xml emd-43129.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43129_fsc.xml emd_43129_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_43129.png emd_43129.png | 141.6 KB | ||
| Filedesc metadata |  emd-43129.cif.gz emd-43129.cif.gz | 6.8 KB | ||
| Others |  emd_43129_half_map_1.map.gz emd_43129_half_map_1.map.gz emd_43129_half_map_2.map.gz emd_43129_half_map_2.map.gz | 86 MB 86 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43129 http://ftp.pdbj.org/pub/emdb/structures/EMD-43129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43129 | HTTPS FTP |
-Related structure data
| Related structure data |  8vc1MC  8vc2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43129.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43129.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of unbound BmGr9 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A of unbound BmGr9
| File | emd_43129_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of unbound BmGr9 | ||||||||||||
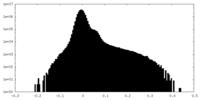
| Projections & Slices |
| ||||||||||||
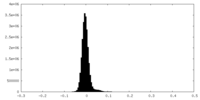
| Density Histograms |
-Half map: Half map B of unbound BmGr9
| File | emd_43129_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of unbound BmGr9 | ||||||||||||
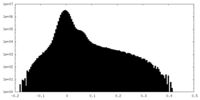
| Projections & Slices |
| ||||||||||||
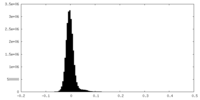
| Density Histograms |
- Sample components
Sample components
-Entire : Agonist-free BmGr9 homotetramer
| Entire | Name: Agonist-free BmGr9 homotetramer |
|---|---|
| Components |
|
-Supramolecule #1: Agonist-free BmGr9 homotetramer
| Supramolecule | Name: Agonist-free BmGr9 homotetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 218.05 KDa |
-Macromolecule #1: Gustatory receptor
| Macromolecule | Name: Gustatory receptor / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.57232 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSHPQFEK GGGSGGGSGG SAWSHPQFEK AALEVLFQGP GTMPPSPDLR ADEPKTPCLV GGAHAFILKI SSFCGLAPLR FEPRSQEYA VTISKGKCFY SYILVTFLVI CTIYGLVAEI GVGVEKSVRM SSRMSQVVSA CDILVVAVTA GVGVYGAPAR M RTMLSYME ...String: MGWSHPQFEK GGGSGGGSGG SAWSHPQFEK AALEVLFQGP GTMPPSPDLR ADEPKTPCLV GGAHAFILKI SSFCGLAPLR FEPRSQEYA VTISKGKCFY SYILVTFLVI CTIYGLVAEI GVGVEKSVRM SSRMSQVVSA CDILVVAVTA GVGVYGAPAR M RTMLSYME NIVAVDRELG RHHSAATERK LCALLLLILL SFTILLVDDF CFYAMQAGKT GRQWEIVTNY AGFYFLWYIV MV LELQFAF TALSLRARLK LFNEALNVTA SQVCKPVKKP KNSQLSVYAT SVRPVSCKRE NVIVETIRVR DKDDAFVMMK TAD GVPCLQ VPPCEAVGRL SRMRCTLCEV TRHIADGYGL PLVIILMSTL LHLIVTPYFL IMEIIVSTHR LHFLVLQFLW CTTH LIRML VVVEPCHYTM REGKRTEDIV CRLMTLAPHG GVLSSKLEVL SRLLMLQNIS YSPLGMCTLD RPLIVTVLGA VTTYL VILI QFQRYDS UniProtKB: Gustatory receptor |
-Macromolecule #2: (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)MET...
| Macromolecule | Name: (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE type: ligand / ID: 2 / Number of copies: 8 / Formula: PSC |
|---|---|
| Molecular weight | Theoretical: 759.068 Da |
| Chemical component information |  ChemComp-PSC: |
-Macromolecule #3: (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5...
| Macromolecule | Name: (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE type: ligand / ID: 3 / Number of copies: 20 / Formula: PC7 |
|---|---|
| Molecular weight | Theoretical: 763.1 Da |
| Chemical component information |  ChemComp-PC7: |
-Macromolecule #4: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 4 / Number of copies: 4 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.25 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11474 / Average exposure time: 3.803 sec. / Average electron dose: 74.492 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 60240 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: ColabFold generated |
|---|---|
| Details | Initial fitting was done using DockInMap in PHENIX and refined through cycles of manual rebuilding in Coot, real-space refinement in PHENIX, and remodeling by simulations run in the ISOLDE plugin of ChimeraX. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 58.19 / Target criteria: cross-correlation coefficient |
| Output model |  PDB-8vc1: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)