[English] 日本語
 Yorodumi
Yorodumi- EMDB-43081: Cryo-EM Structure of a Proteolytic ClpXP AAA+ Machine Poised to U... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of a Proteolytic ClpXP AAA+ Machine Poised to Unfold a Branched-Degron DHFR-ssrA Substrate Bound with MTX | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ClpXP / full-engaged state / AAA protease / CHAPERONE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein denaturation / HslUV protease complex / endopeptidase Clp / endopeptidase Clp complex / positive regulation of programmed cell death / NADP+ binding / response to temperature stimulus / ATP-dependent peptidase activity / dihydrofolate metabolic process / dihydrofolate reductase ...protein denaturation / HslUV protease complex / endopeptidase Clp / endopeptidase Clp complex / positive regulation of programmed cell death / NADP+ binding / response to temperature stimulus / ATP-dependent peptidase activity / dihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / protein quality control for misfolded or incompletely synthesized proteins / protein unfolding / tetrahydrofolate biosynthetic process / proteasomal protein catabolic process / one-carbon metabolic process / serine-type peptidase activity / : / ATP-dependent protein folding chaperone / response to radiation / disordered domain specific binding / unfolded protein binding / response to heat / ATPase binding / protease binding / protein dimerization activity / serine-type endopeptidase activity / cell division / ATP hydrolysis activity / proteolysis / zinc ion binding / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||
 Authors Authors | Ghanbarpour A / Sauer RT / Davis JH | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A proteolytic AAA+ machine poised to unfold protein substrates. Authors: Alireza Ghanbarpour / Robert T Sauer / Joseph H Davis /  Abstract: AAA+ proteolytic machines unfold proteins before degrading them. Here, we present cryoEM structures of ClpXP-substrate complexes that reveal a postulated but heretofore unseen intermediate in ...AAA+ proteolytic machines unfold proteins before degrading them. Here, we present cryoEM structures of ClpXP-substrate complexes that reveal a postulated but heretofore unseen intermediate in substrate unfolding/degradation. A ClpX hexamer draws natively folded substrates tightly against its axial channel via interactions with a fused C-terminal degron tail and ClpX-RKH loops that flexibly conform to the globular substrate. The specific ClpX-substrate contacts observed vary depending on the substrate degron and affinity tags, helping to explain ClpXP's ability to unfold/degrade a wide array of different cellular substrates. Some ClpX contacts with native substrates are enabled by upward movement of the seam subunit in the AAA+ spiral, a motion coupled to a rearrangement of contacts between the ClpX unfoldase and ClpP peptidase. Our structures additionally highlight ClpX's ability to translocate a diverse array of substrate topologies, including the co-translocation of two polypeptide chains. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43081.map.gz emd_43081.map.gz | 55 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43081-v30.xml emd-43081-v30.xml emd-43081.xml emd-43081.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43081_fsc.xml emd_43081_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_43081.png emd_43081.png | 58.9 KB | ||
| Filedesc metadata |  emd-43081.cif.gz emd-43081.cif.gz | 6.8 KB | ||
| Others |  emd_43081_half_map_1.map.gz emd_43081_half_map_1.map.gz emd_43081_half_map_2.map.gz emd_43081_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43081 http://ftp.pdbj.org/pub/emdb/structures/EMD-43081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43081 | HTTPS FTP |
-Related structure data
| Related structure data |  8v9rMC  9c87C  9c88C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43081.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43081.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1245 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ClpX.ClpP.DHFR
| Entire | Name: ClpX.ClpP.DHFR |
|---|---|
| Components |
|
-Supramolecule #1: ClpX.ClpP.DHFR
| Supramolecule | Name: ClpX.ClpP.DHFR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent Clp protease ATP-binding subunit ClpX
| Macromolecule | Name: ATP-dependent Clp protease ATP-binding subunit ClpX / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.355812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH DYDIPTTENL YFQGSSALPT PHEIRNHLDD YVIGQEQAKK VLAVAVYNHY KRLRNGDTSN GVELGKSNIL LIGPTGSGK TLLAETLARL LDVPFTMADA TTLTEAGYVG EDVENIIQKL LQKSDYDVQK AQRGIVYIDE IDKISRKSDN P SITRDVSG ...String: MGSSHHHHHH DYDIPTTENL YFQGSSALPT PHEIRNHLDD YVIGQEQAKK VLAVAVYNHY KRLRNGDTSN GVELGKSNIL LIGPTGSGK TLLAETLARL LDVPFTMADA TTLTEAGYVG EDVENIIQKL LQKSDYDVQK AQRGIVYIDE IDKISRKSDN P SITRDVSG EGVQQALLKL IEGTVAAVPP QGGRKHPQQE FLQVDTSKIL FICGGAFAGL DKVISHRVET GSGIGFGATV KA KSDKASE GELLAQVEPE DLIKFGLIPE FIGRLPVVAT LNELSEEALI QILKEPKNAL TKQYQALFNL EGVDLEFRDE ALD AIAKKA MARKTGARGL RSIVEAALLD TMYDLPSMED VEKVVIDESV IDGQSEPLLI YGKPEAQQAS GE UniProtKB: ATP-dependent Clp protease ATP-binding subunit ClpX |
-Macromolecule #2: Dihydrofolate reductase
| Macromolecule | Name: Dihydrofolate reductase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.658607 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MISLIAALAV DRVIGMENAM PWNLPADLAW FKRNTLDKPV IMGRHTWESI GRPLPGRKNI ILSSQPGTDD RVTWVKSVDE AIAACGDVP EIMVIGGGRV YEQFLPKAQK LYLTHIDAEV EGDTHFPDYE PDDWESVFSE FHDADAQNSH SYCFEILERR G SHLGLIEV ...String: MISLIAALAV DRVIGMENAM PWNLPADLAW FKRNTLDKPV IMGRHTWESI GRPLPGRKNI ILSSQPGTDD RVTWVKSVDE AIAACGDVP EIMVIGGGRV YEQFLPKAQK LYLTHIDAEV EGDTHFPDYE PDDWESVFSE FHDADAQNSH SYCFEILERR G SHLGLIEV EKPLYCVEPF VGETAHFEIE LSEPDVHGQW KLTSHHHHHH UniProtKB: dihydrofolate reductase |
-Macromolecule #3: ATP-dependent Clp protease proteolytic subunit
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit / type: protein_or_peptide / ID: 3 / Number of copies: 14 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.468869 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LVPMVIEQTS RGERSFDIYS RLLKERVIFL TGQVEDHMAN LIVAQMLFLE AENPEKDIYL YINSPGGVIT AGMSIYDTMQ FIKPDVSTI CMGQAASMGA FLLTAGAKGK RFCLPNSRVM IHQPLGGYQG QATDIEIHAR EILKVKGRMN ELMALHTGQS L EQIERDTE ...String: LVPMVIEQTS RGERSFDIYS RLLKERVIFL TGQVEDHMAN LIVAQMLFLE AENPEKDIYL YINSPGGVIT AGMSIYDTMQ FIKPDVSTI CMGQAASMGA FLLTAGAKGK RFCLPNSRVM IHQPLGGYQG QATDIEIHAR EILKVKGRMN ELMALHTGQS L EQIERDTE RDRFLSAPEA VEYGLVDSIL THRNENLYFQ SLEHHHHHH UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #7: METHOTREXATE
| Macromolecule | Name: METHOTREXATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: MTX |
|---|---|
| Molecular weight | Theoretical: 454.439 Da |
| Chemical component information | 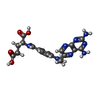 ChemComp-MTX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.94 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































