[English] 日本語
 Yorodumi
Yorodumi- EMDB-42890: Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acy... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | herpes simplex virus / replication / DNA polymerase / ACYCLOVIR / antiherpesvirus drug / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationbidirectional double-stranded viral DNA replication / exonuclease activity / DNA-templated DNA replication / RNA-DNA hybrid ribonuclease activity / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / host cell nucleus / DNA binding Similarity search - Function | |||||||||
| Biological species |   Human alphaherpesvirus 1 strain KOS / synthetic construct (others) Human alphaherpesvirus 1 strain KOS / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Pan J / Abraham J / Coen DM / Shankar S / Yang P / Hogle J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance. Authors: Sundaresh Shankar / Junhua Pan / Pan Yang / Yuemin Bian / Gábor Oroszlán / Zishuo Yu / Purba Mukherjee / David J Filman / James M Hogle / Mrinal Shekhar / Donald M Coen / Jonathan Abraham /    Abstract: DNA polymerases are important drug targets, and many structural studies have captured them in distinct conformations. However, a detailed understanding of the impact of polymerase conformational ...DNA polymerases are important drug targets, and many structural studies have captured them in distinct conformations. However, a detailed understanding of the impact of polymerase conformational dynamics on drug resistance is lacking. We determined cryoelectron microscopy (cryo-EM) structures of DNA-bound herpes simplex virus polymerase holoenzyme in multiple conformations and interacting with antivirals in clinical use. These structures reveal how the catalytic subunit Pol and the processivity factor UL42 bind DNA to promote processive DNA synthesis. Unexpectedly, in the absence of an incoming nucleotide, we observed Pol in multiple conformations with the closed state sampled by the fingers domain. Drug-bound structures reveal how antivirals may selectively bind enzymes that more readily adopt the closed conformation. Molecular dynamics simulations and the cryo-EM structure of a drug-resistant mutant indicate that some resistance mutations modulate conformational dynamics rather than directly impacting drug binding, thus clarifying mechanisms that drive drug selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42890.map.gz emd_42890.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42890-v30.xml emd-42890-v30.xml emd-42890.xml emd-42890.xml | 30 KB 30 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42890_fsc.xml emd_42890_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_42890.png emd_42890.png | 76.5 KB | ||
| Filedesc metadata |  emd-42890.cif.gz emd-42890.cif.gz | 8.5 KB | ||
| Others |  emd_42890_additional_1.map.gz emd_42890_additional_1.map.gz emd_42890_half_map_1.map.gz emd_42890_half_map_1.map.gz emd_42890_half_map_2.map.gz emd_42890_half_map_2.map.gz | 98 MB 98.6 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42890 http://ftp.pdbj.org/pub/emdb/structures/EMD-42890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42890 | HTTPS FTP |
-Validation report
| Summary document |  emd_42890_validation.pdf.gz emd_42890_validation.pdf.gz | 954 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42890_full_validation.pdf.gz emd_42890_full_validation.pdf.gz | 953.6 KB | Display | |
| Data in XML |  emd_42890_validation.xml.gz emd_42890_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_42890_validation.cif.gz emd_42890_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42890 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42890 | HTTPS FTP |
-Related structure data
| Related structure data |  8v1tMC  8exxC  8v1qC  8v1rC  8v1sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : HERPES SIMPLEX VIRUS TYPE 1 POLYMERASE HOLOENZYME UL30:UL42 IN CO...
| Entire | Name: HERPES SIMPLEX VIRUS TYPE 1 POLYMERASE HOLOENZYME UL30:UL42 IN COMPLEX WITH dsDNA AND ACYCLOVIR TRIPHOSPHATE |
|---|---|
| Components |
|
-Supramolecule #1: HERPES SIMPLEX VIRUS TYPE 1 POLYMERASE HOLOENZYME UL30:UL42 IN CO...
| Supramolecule | Name: HERPES SIMPLEX VIRUS TYPE 1 POLYMERASE HOLOENZYME UL30:UL42 IN COMPLEX WITH dsDNA AND ACYCLOVIR TRIPHOSPHATE type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: EACH COMPLEX CONSISTS OF ONE HSV-1 UL30 (expressed in insect cells), ONE HSV-1 UL42 (expressed in E.coli), ONE TEMPLATE DNA (synthetic), ONE PRIMER DNA (synthetic), AND ONE ACYCLOVIR ...Details: EACH COMPLEX CONSISTS OF ONE HSV-1 UL30 (expressed in insect cells), ONE HSV-1 UL42 (expressed in E.coli), ONE TEMPLATE DNA (synthetic), ONE PRIMER DNA (synthetic), AND ONE ACYCLOVIR TRIPHOSPHATE MOLECULE (synthetic). |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS / Strain: KOS Human alphaherpesvirus 1 strain KOS / Strain: KOS |
| Molecular weight | Theoretical: 197 KDa |
-Macromolecule #1: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 1 Details: Herpes simplex virus type 1 (KOS strain) DNA polymerase catalytic subunit UL30 with its N-terminal 42 residues deleted and replaced by an N-terminal poly-histidine tag in the expression construct Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS Human alphaherpesvirus 1 strain KOS |
| Molecular weight | Theoretical: 133.614344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHNFYN PYLAPVGTQQ KPTGPTQRHT YYSECDEFRF IAPRVLDEDA PPEKRAGVHD GHLKRAPKVY CGGDERDVLR VGSGGFWPR RSRLWGGVDH APAGFNPTVT VFHVYDILEN VEHAYGMRAA QFHARFMDAI TPTGTVITLL GLTPEGHRVA V HVYGTRQY ...String: HHHHHHNFYN PYLAPVGTQQ KPTGPTQRHT YYSECDEFRF IAPRVLDEDA PPEKRAGVHD GHLKRAPKVY CGGDERDVLR VGSGGFWPR RSRLWGGVDH APAGFNPTVT VFHVYDILEN VEHAYGMRAA QFHARFMDAI TPTGTVITLL GLTPEGHRVA V HVYGTRQY FYMNKEEVDR HLQCRAPRDL CERMAAALRE SPGASFRGIS ADHFEAEVVE RTDVYYYETR PALFYRVYVR SG RVLSYLC DNFCPAIKKY EGGVDATTRF ILDNPGFVTF GWYRLKPGRN NTLAQPRAPM AFGTSSDVEF NCTADNLAIE GGM SDLPAY KLMCFDIECK AGGEDELAFP VAGHPEDLVI QISCLLYDLS TTALEHVLLF SLGSCDLPES HLNELAARGL PTPV VLEFD SEFEMLLAFM TLVKQYGPEF VTGYNIINFD WPFLLAKLTD IYKVPLDGYG RMNGRGVFRV WDIGQSHFQK RSKIK VNGM VNIDMYGIIT DKIKLSSYKL NAVAEAVLKD KKKDLSYRDI PAYYATGPAQ RGVIGEYCIQ DSLLVGQLFF KFLPHL ELS AVARLAGINI TRTIYDGQQI RVFTCLLRLA DQKGFILPDT QGRFRGAGGE APKRPAAARE DEERPEEEGE DEDEREE GG GEREPEGARE TAGRHVGYQG ARVLDPTSGF HVNPVVVFDF ASLYPSIIQA HNLCFSTLSL RADAVAHLEA GKDYLEIE V GGRRLFFVKA HVRESLLSIL LRDWLAMRKQ IRSRIPQSSP EEAVLLDKQQ AAIKVVCNSV YGFTGVQHGL LPCLHVAAT VTTIGREMLL ATREYVHARW AAFEQLLADF PEAADMRAPG PYSMRIIYGD TDSIFVLCRG LTAAGLTAMG DKMASHISRA LFLPPIKLE CEKTFTKLLL IAKKKYIGVI YGGKMLIKGV DLVRKNNCAF INRTSRALVD LLFYDDTVSG AAAALAERPA E EWLARPLP EGLQAFGAVL VDAHRRITDP ERDIQDFVLT AELSRHPRAY TNKRLAHLTV YYKLMARRAQ VPSIKDRIPY VI VAQTREV EETVARLAAL RELDAAAPGD EPAPPAALPS PAKRPRETPS HADPPGGASK PRKLLVSELA EDPAYAIAHG VAL NTDYYF SHLLGAACVT FKALFGNNAK ITESLLKRFI PEVWHPPDDV AARLRAAGFG AVGAGATAEE TRRMLHRAFD TLA UniProtKB: DNA polymerase |
-Macromolecule #2: DNA polymerase processivity factor
| Macromolecule | Name: DNA polymerase processivity factor / type: protein_or_peptide / ID: 2 Details: Herpes simplex virus 1 (KOS strain) DNA polymerase processivity factor UL42 residues 1-340 tagged with a prescission protease cleavage site followed by a maltose binding proten (MBP) tag in ...Details: Herpes simplex virus 1 (KOS strain) DNA polymerase processivity factor UL42 residues 1-340 tagged with a prescission protease cleavage site followed by a maltose binding proten (MBP) tag in the expression construct Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS Human alphaherpesvirus 1 strain KOS |
| Molecular weight | Theoretical: 36.34618 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDSPGGVAP ASPVEDASDA SLGQPEEGAP CQVVLQGAEL NGILQAFAPL RTSLLDSLLV MGDRGILIHN TIFGEQVFLP LEHSQFSRY RWRGPTAAFL SLVDQKRSLL SVFRANQYPD LRRVELAITG QAPFRTLVQR IWTTTSDGEA VELASETLMK R ELTSFVVL ...String: MTDSPGGVAP ASPVEDASDA SLGQPEEGAP CQVVLQGAEL NGILQAFAPL RTSLLDSLLV MGDRGILIHN TIFGEQVFLP LEHSQFSRY RWRGPTAAFL SLVDQKRSLL SVFRANQYPD LRRVELAITG QAPFRTLVQR IWTTTSDGEA VELASETLMK R ELTSFVVL VPQGTPDVQL RLTRPQLTKV LNATGADSAT PTTFELGVNG KFSVFTTSTC VTFAAREEGV SSSTSTQVQI LS NALTKAG QAAANAKTVY GENTHRTFSV VVDDCSMRAV LRRLQVAGGT LKFFLTTPVP SLCVTATGPN AVSAVFLLKP QKI CLDWLG HSQGSPSAGS SASR UniProtKB: DNA polymerase processivity factor |
-Macromolecule #3: PRIMER DNA (32-MER)
| Macromolecule | Name: PRIMER DNA (32-MER) / type: dna / ID: 3 Details: 3'-deoxyl primer DNA strand, no mismatch with template strand Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.866353 KDa |
| Sequence | String: (DG)(DA)(DT)(DT)(DA)(DC)(DG)(DA)(DA)(DT) (DT)(DC)(DG)(DA)(DG)(DC)(DT)(DC)(DG)(DG) (DT)(DA)(DC)(DC)(DC)(DG)(DG)(DG)(DG) (DA)(DT)(DOC) |
-Macromolecule #4: TEMPLATE DNA (50-MER)
| Macromolecule | Name: TEMPLATE DNA (50-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.199773 KDa |
| Sequence | String: (DC)(DA)(DC)(DA)(DC)(DA)(DC)(DA)(DC)(DA) (DC)(DA)(DC)(DA)(DC)(DA)(DC)(DC)(DG)(DA) (DT)(DC)(DC)(DC)(DC)(DG)(DG)(DG)(DT) (DA)(DC)(DC)(DG)(DA)(DG)(DC)(DT)(DC)(DG) (DA) (DA)(DT)(DT)(DC)(DG)(DT)(DA)(DA) (DT)(DC) |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ACYCLOVIR TRIPHOSPHATE
| Macromolecule | Name: ACYCLOVIR TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: AVP |
|---|---|
| Molecular weight | Theoretical: 465.144 Da |
| Chemical component information | 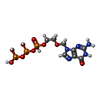 ChemComp-AVP: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.38 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM tris(2-carboxyethyl)phosphine (TCEP) |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: GRIDS WERE GLOW DISCHARGED IN A PELCO EASIGLOW AT 15 MA FOR 30 SECONDS UNDER 0.39 MBAR. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 3 MICROLITERS OF SAMPLE WERE BLOTTED FOR 3 SECONDS WITH FILTER PAPER SATURATED UNDER 100% HUMIDITY PRIOR TO PLUNGING.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Software | Name: SerialEM (ver. 3.8) |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 3268 / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 60606 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  UCSF Chimera (ver. 1.13.1) UCSF Chimera (ver. 1.13.1) |
|---|---|
| Details | RIGID BODY, MINIMIZATION_GLOBAL, LOCAL_GRID_SEARCH, ADP REFINEMENT |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 33.64 / Target criteria: CORRELATION COEFFICIENT |
| Output model |  PDB-8v1t: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















