[English] 日本語
 Yorodumi
Yorodumi- EMDB-42590: Prototypic SARS-CoV-2 spike (containing K417) in the open conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | |||||||||
 Map data Map data | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Prototypic SARS-CoV-2 spike / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Geng Q / Liu B / Li F | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein. Authors: Qibin Geng / Yushun Wan / Fu-Chun Hsueh / Jian Shang / Gang Ye / Fan Bu / Morgan Herbst / Rowan Wilkens / Bin Liu / Fang Li /  Abstract: SARS-CoV-2 spike protein plays a key role in mediating viral entry and inducing host immune responses. It can adopt either an open or closed conformation based on the position of its receptor-binding ...SARS-CoV-2 spike protein plays a key role in mediating viral entry and inducing host immune responses. It can adopt either an open or closed conformation based on the position of its receptor-binding domain (RBD). It is yet unclear what causes these conformational changes or how they influence the spike's functions. Here, we show that Lys417 in the RBD plays dual roles in the spike's structure: it stabilizes the closed conformation of the trimeric spike by mediating inter-spike-subunit interactions; it also directly interacts with ACE2 receptor. Hence, a K417V mutation has opposing effects on the spike's function: it opens up the spike for better ACE2 binding while weakening the RBD's direct binding to ACE2. The net outcomes of this mutation are to allow the spike to bind ACE2 with higher probability and mediate viral entry more efficiently, but become more exposed to neutralizing antibodies. Given that residue 417 has been a viral mutational hotspot, SARS-CoV-2 may have been evolving to strike a balance between infection potency and immune evasion, contributing to its pandemic spread. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42590.map.gz emd_42590.map.gz | 107.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42590-v30.xml emd-42590-v30.xml emd-42590.xml emd-42590.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42590_fsc.xml emd_42590_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42590.png emd_42590.png | 83 KB | ||
| Filedesc metadata |  emd-42590.cif.gz emd-42590.cif.gz | 7.1 KB | ||
| Others |  emd_42590_additional_1.map.gz emd_42590_additional_1.map.gz emd_42590_half_map_1.map.gz emd_42590_half_map_1.map.gz emd_42590_half_map_2.map.gz emd_42590_half_map_2.map.gz | 204 MB 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42590 http://ftp.pdbj.org/pub/emdb/structures/EMD-42590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42590 | HTTPS FTP |
-Validation report
| Summary document |  emd_42590_validation.pdf.gz emd_42590_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42590_full_validation.pdf.gz emd_42590_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42590_validation.xml.gz emd_42590_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_42590_validation.cif.gz emd_42590_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42590 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42590 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42590 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42590 | HTTPS FTP |
-Related structure data
| Related structure data |  8uumMC  8uulC  8uunC  8uuoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42590.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42590.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
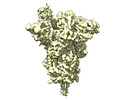
| Annotation | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88533 Å | ||||||||||||||||||||||||||||||||||||
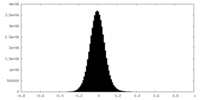
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Prototypic SARS-CoV-2 spike (containing K417) in the open...
| File | emd_42590_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
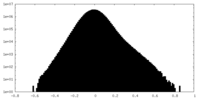
| Density Histograms |
-Half map: Prototypic SARS-CoV-2 spike (containing K417) in the open...
| File | emd_42590_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation -half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Prototypic SARS-CoV-2 spike (containing K417) in the open...
| File | emd_42590_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation -half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Prototypic SARS-CoV-2 spike (containing K417) in the open conformation
| Entire | Name: Prototypic SARS-CoV-2 spike (containing K417) in the open conformation |
|---|---|
| Components |
|
-Supramolecule #1: Prototypic SARS-CoV-2 spike (containing K417) in the open conformation
| Supramolecule | Name: Prototypic SARS-CoV-2 spike (containing K417) in the open conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 411 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.140781 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPA GAGSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQYIK UniProtKB: Spike glycoprotein |
-Macromolecule #3: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 3776 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)