+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacillus niacini flavin monooxygenase with bound (2,6)DHP | |||||||||
 Map data Map data | Primary map filtered by estimated local resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Monooxygenase / flavin binding / CYTOSOLIC PROTEIN | |||||||||
| Biological species |  Neobacillus niacini (bacteria) Neobacillus niacini (bacteria) | |||||||||
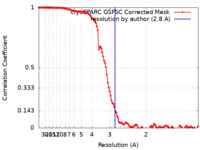
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Richardson BC / French JB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2024 Journal: Biochemistry / Year: 2024Title: Structural and Functional Characterization of a Novel Class A Flavin Monooxygenase from . Authors: Brian C Richardson / Zachary R Turlington / Sofia Vaz Ferreira de Macedo / Sara K Phillips / Kay Perry / Savannah G Brancato / Emmalee W Cooke / Jonathan R Gwilt / Morgan A Dasovich / Andrew ...Authors: Brian C Richardson / Zachary R Turlington / Sofia Vaz Ferreira de Macedo / Sara K Phillips / Kay Perry / Savannah G Brancato / Emmalee W Cooke / Jonathan R Gwilt / Morgan A Dasovich / Andrew J Roering / Francis M Rossi / Mark J Snider / Jarrod B French / Katherine A Hicks /  Abstract: A gene cluster responsible for the degradation of nicotinic acid (NA) in has recently been identified, and the structures and functions of the resulting enzymes are currently being evaluated to ...A gene cluster responsible for the degradation of nicotinic acid (NA) in has recently been identified, and the structures and functions of the resulting enzymes are currently being evaluated to establish pathway intermediates. One of the genes within this cluster encodes a flavin monooxygenase (BnFMO) that is hypothesized to catalyze a hydroxylation reaction. Kinetic analyses of the recombinantly purified BnFMO suggest that this enzyme catalyzes the hydroxylation of 2,6-dihydroxynicotinic acid (2,6-DHNA) or 2,6-dihydroxypyridine (2,6-DHP), which is formed spontaneously by the decarboxylation of 2,6-DHNA. To understand the details of this hydroxylation reaction, we determined the structure of BnFMO using a multimodel approach combining protein X-ray crystallography and cryo-electron microscopy (cryo-EM). A liganded BnFMO cryo-EM structure was obtained in the presence of 2,6-DHP, allowing us to make predictions about potential catalytic residues. The structural data demonstrate that BnFMO is trimeric, which is unusual for Class A flavin monooxygenases. In both the electron density and coulomb potential maps, a region at the trimeric interface was observed that was consistent with and modeled as lipid molecules. High-resolution mass spectral analysis suggests that there is a mixture of phosphatidylethanolamine and phosphatidylglycerol lipids present. Together, these data provide insights into the molecular details of the central hydroxylation reaction unique to the aerobic degradation of NA in . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42490.map.gz emd_42490.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42490-v30.xml emd-42490-v30.xml emd-42490.xml emd-42490.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42490_fsc.xml emd_42490_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42490.png emd_42490.png | 107.1 KB | ||
| Filedesc metadata |  emd-42490.cif.gz emd-42490.cif.gz | 6.9 KB | ||
| Others |  emd_42490_additional_1.map.gz emd_42490_additional_1.map.gz emd_42490_half_map_1.map.gz emd_42490_half_map_1.map.gz emd_42490_half_map_2.map.gz emd_42490_half_map_2.map.gz | 82.5 MB 154.3 MB 154.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42490 http://ftp.pdbj.org/pub/emdb/structures/EMD-42490 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42490 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42490 | HTTPS FTP |
-Related structure data
| Related structure data |  8urdMC  8uiuC  8urcC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42490.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42490.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map filtered by estimated local resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.665 Å | ||||||||||||||||||||||||||||||||||||
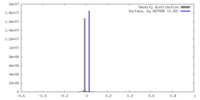
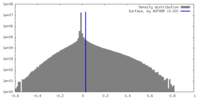
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : flavin monooxygenase asymmetric trimer
| Entire | Name: flavin monooxygenase asymmetric trimer |
|---|---|
| Components |
|
-Supramolecule #1: flavin monooxygenase asymmetric trimer
| Supramolecule | Name: flavin monooxygenase asymmetric trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Neobacillus niacini (bacteria) Neobacillus niacini (bacteria) |
-Macromolecule #1: Flavin monooxygenase
| Macromolecule | Name: Flavin monooxygenase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neobacillus niacini (bacteria) Neobacillus niacini (bacteria) |
| Molecular weight | Theoretical: 51.924461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGENLYFQG HMEELIKEVQ SDVCIVGAGP AGMLLGLLLA KQGLEVIVLE QNGDFHREYR GEITQPRFVQ LMKQLNLLD YIESNSHVKI PEVNVFHNNV KIMQLAFNTL IDEESYCARL TQPTLLSALL DKAKKYPNFK LLFNTKVRDL L REDGKVTG ...String: MGSSHHHHHH SSGENLYFQG HMEELIKEVQ SDVCIVGAGP AGMLLGLLLA KQGLEVIVLE QNGDFHREYR GEITQPRFVQ LMKQLNLLD YIESNSHVKI PEVNVFHNNV KIMQLAFNTL IDEESYCARL TQPTLLSALL DKAKKYPNFK LLFNTKVRDL L REDGKVTG VYAVAKPGEQ INFTEDEVFE GNLNIKSRVT VGVDGRNSTM EKLGNFELEL DYYDNDLLWF SFEKPESWDY NI YHFYFQK NYNYLFLPKL GGYIQCGISL TKGEYQKIKK EGIESFKEKI LEDMPILKQH FDTVTDFKSF VQLLCRMRYI KDW AKEEGC MLIGDAAHCV TPWGAVGSTL AMGTAVIAAD VIYKGFKNND LSLETLKQVQ SRRKEEVKMI QNLQLTIEKF LTRE PIKKE IAPLMFSIAT KMPDITNLYK KLFTREFPLD IDESFIFHDE LVEAN |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 3 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: pyridine-2,6-diol
| Macromolecule | Name: pyridine-2,6-diol / type: ligand / ID: 3 / Number of copies: 3 / Formula: WTQ |
|---|---|
| Molecular weight | Theoretical: 111.099 Da |
-Macromolecule #4: 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL
| Macromolecule | Name: 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL type: ligand / ID: 4 / Number of copies: 3 / Formula: DR9 |
|---|---|
| Molecular weight | Theoretical: 746.991 Da |
| Chemical component information |  ChemComp-DR9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM Tris pH 8 50 mM NaCl | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 7048 / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)