[English] 日本語
 Yorodumi
Yorodumi- EMDB-42487: Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer of dimers | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oleate hydratase (OhyA) / phospholipids / membrane binding domain / amphipathic helices / interfacial enzyme / peripheral membrane protein / HYDROLASE | |||||||||
| Function / homology | oleate hydratase / oleate hydratase activity / Oleate hydratase / MCRA family / FAD binding / fatty acid metabolic process / FAD/NAD(P)-binding domain superfamily / Myosin-cross-reactive antigen Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
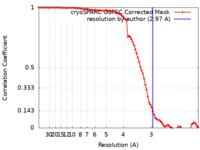
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Oldham ML / Qayyum MZ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2024 Journal: J Struct Biol / Year: 2024Title: Cryo-EM reconstruction of oleate hydratase bound to a phospholipid membrane bilayer. Authors: Michael L Oldham / M Zuhaib Qayyum / Ravi C Kalathur / Charles O Rock / Christopher D Radka /  Abstract: Oleate hydratase (OhyA) is a bacterial peripheral membrane protein that catalyzes FAD-dependent water addition to membrane bilayer-embedded unsaturated fatty acids. The opportunistic pathogen ...Oleate hydratase (OhyA) is a bacterial peripheral membrane protein that catalyzes FAD-dependent water addition to membrane bilayer-embedded unsaturated fatty acids. The opportunistic pathogen Staphylococcus aureus uses OhyA to counteract the innate immune system and support colonization. Many Gram-positive and Gram-negative bacteria in the microbiome also encode OhyA. OhyA is a dimeric flavoenzyme whose carboxy terminus is identified as the membrane binding domain; however, understanding how OhyA binds to cellular membranes is not complete until the membrane-bound structure has been elucidated. All available OhyA structures depict the solution state of the protein outside its functional environment. Here, we employ liposomes to solve the cryo-electron microscopy structure of the functional unit: the OhyA•membrane complex. The protein maintains its structure upon membrane binding and slightly alters the curvature of the liposome surface. OhyA preferentially associates with 20-30 nm liposomes with multiple copies of OhyA dimers assembling on the liposome surface resulting in the formation of higher-order oligomers. Dimer assembly is cooperative and extends along a formed ridge of the liposome. We also solved an OhyA dimer of dimers structure that recapitulates the intermolecular interactions that stabilize the dimer assembly on the membrane bilayer as well as the crystal contacts in the lattice of the OhyA crystal structure. Our work enables visualization of the molecular trajectory of membrane binding for this important interfacial enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42487.map.gz emd_42487.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42487-v30.xml emd-42487-v30.xml emd-42487.xml emd-42487.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42487_fsc.xml emd_42487_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42487.png emd_42487.png | 67.5 KB | ||
| Filedesc metadata |  emd-42487.cif.gz emd-42487.cif.gz | 6.6 KB | ||
| Others |  emd_42487_additional_1.map.gz emd_42487_additional_1.map.gz emd_42487_half_map_1.map.gz emd_42487_half_map_1.map.gz emd_42487_half_map_2.map.gz emd_42487_half_map_2.map.gz | 59.8 MB 62.2 MB 62.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42487 http://ftp.pdbj.org/pub/emdb/structures/EMD-42487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42487 | HTTPS FTP |
-Related structure data
| Related structure data |  8ur8M M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42487.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42487.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.044 Å | ||||||||||||||||||||||||||||||||||||
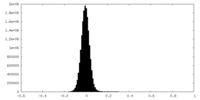
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer map
| File | emd_42487_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
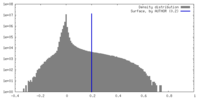
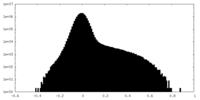
| Density Histograms |
-Half map: #2
| File | emd_42487_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
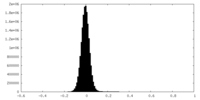
| Density Histograms |
-Half map: #1
| File | emd_42487_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
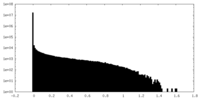
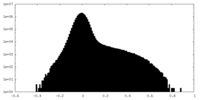
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of Dimers of OhyA
| Entire | Name: Dimer of Dimers of OhyA |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of Dimers of OhyA
| Supramolecule | Name: Dimer of Dimers of OhyA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 278.686 KDa |
-Macromolecule #1: Oleate hydratase
| Macromolecule | Name: Oleate hydratase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.892719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MYYSYGNYEA FARPKKPENV ENKSAYLIGS GLASLAAACF LIRDGQMEGS KIHILEELPK AGGSLDGEN MPLKGYVVRG GREMENHFEC LWDLFRSIPS LEIDNASVLD EFYWLNKEDP NYSRCRVIEK QGQRLVTDGD F TLTKTAIK ...String: MGSSHHHHHH SSGLVPRGSH MYYSYGNYEA FARPKKPENV ENKSAYLIGS GLASLAAACF LIRDGQMEGS KIHILEELPK AGGSLDGEN MPLKGYVVRG GREMENHFEC LWDLFRSIPS LEIDNASVLD EFYWLNKEDP NYSRCRVIEK QGQRLVTDGD F TLTKTAIK EILDLCLTNE EDLDDVKITD VFSDDFFNSN FWIYWKTMFA FEPWHSAMEM RRYLMRFVHH ISGLADFSAL KF TKYNQYE SLVLPMVEYL KSHGVQFEYD VKVEDIKIDV TTSQKIAREI LIDRNGNAES IKLTINDLVF VTNGSITESS TYG DNDTPA PPTDELGGSW TLWKNLARQS PEFGNPDKFC QNIPKKSWFV SATSTTNNKE IIDTIESICK RDPLAGKTVT GGII TINDS AWQMSFTINR QQQFKDQPEN EISTWIYALY SDVNGDYIKK PITECSGNEI CQEWLYHLGV STDKIEDLAK HASNT IPVY MPYITSYFMT RAIGDRPLVV PHQSQNLAFI GNFAETERDT VFTTEYSVRT AMEAVYQLLN IDRGIPEVIN SPFDLR VLM DAIYELNDHQ DLREITKDSK MQKLALAGFL KKIKGTYIES LLKEHKLL UniProtKB: Myosin-cross-reactive antigen |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.3 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 276 / Average exposure time: 3.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 79000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model / Details: initial model was a crystal structure |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8ur8: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)