[English] 日本語
 Yorodumi
Yorodumi- EMDB-42484: Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oleate hydratase (OhyA) / phospholipids / membrane binding domain / amphipathic helices / interfacial enzyme / peripheral membrane protein / HYDROLASE | |||||||||
| Function / homology | oleate hydratase / oleate hydratase activity / Oleate hydratase / MCRA family / FAD binding / fatty acid metabolic process / FAD/NAD(P)-binding domain superfamily / Myosin-cross-reactive antigen Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
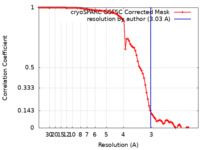
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Oldham ML / Qayyum MZ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: The carboxy terminus causes interfacial assembly of oleate hydratase on a membrane bilayer. Authors: Christopher D Radka / Christy R Grace / Hale S Hasdemir / Yupeng Li / Carlos C Rodriguez / Patrick Rodrigues / Michael L Oldham / M Zuhaib Qayyum / Aaron Pitre / William J MacCain / Ravi C ...Authors: Christopher D Radka / Christy R Grace / Hale S Hasdemir / Yupeng Li / Carlos C Rodriguez / Patrick Rodrigues / Michael L Oldham / M Zuhaib Qayyum / Aaron Pitre / William J MacCain / Ravi C Kalathur / Emad Tajkhorshid / Charles O Rock /  Abstract: The soluble flavoprotein oleate hydratase (OhyA) hydrates the 9-cis double bond of unsaturated fatty acids. OhyA substrates are embedded in membrane bilayers; OhyA must remove the fatty acid from the ...The soluble flavoprotein oleate hydratase (OhyA) hydrates the 9-cis double bond of unsaturated fatty acids. OhyA substrates are embedded in membrane bilayers; OhyA must remove the fatty acid from the bilayer and enclose it in the active site. Here, we show that the positively charged helix-turn-helix motif in the carboxy terminus (CTD) is responsible for interacting with the negatively charged phosphatidylglycerol (PG) bilayer. Super-resolution microscopy of Staphylococcus aureus cells expressing green fluorescent protein fused to OhyA or the CTD sequence shows subcellular localization along the cellular boundary, indicating OhyA is membrane-associated and the CTD sequence is sufficient for membrane recruitment. Using cryo-electron microscopy, we solved the OhyA dimer structure and conducted 3D variability analysis of the reconstructions to assess CTD flexibility. Our surface plasmon resonance experiments corroborated that OhyA binds the PG bilayer with nanomolar affinity and we found the CTD sequence has intrinsic PG binding properties. We determined that the nuclear magnetic resonance structure of a peptide containing the CTD sequence resembles the OhyA crystal structure. We observed intermolecular NOE from PG liposome protons next to the phosphate group to the CTD peptide. The addition of paramagnetic MnCl indicated the CTD peptide binds the PG surface but does not insert into the bilayer. Molecular dynamics simulations, supported by site-directed mutagenesis experiments, identify key residues in the helix-turn-helix that drive membrane association. The data show that the OhyA CTD binds the phosphate layer of the PG surface to obtain bilayer-embedded unsaturated fatty acids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42484.map.gz emd_42484.map.gz | 33.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42484-v30.xml emd-42484-v30.xml emd-42484.xml emd-42484.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42484_fsc.xml emd_42484_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42484.png emd_42484.png | 93 KB | ||
| Filedesc metadata |  emd-42484.cif.gz emd-42484.cif.gz | 6.8 KB | ||
| Others |  emd_42484_additional_1.map.gz emd_42484_additional_1.map.gz emd_42484_half_map_1.map.gz emd_42484_half_map_1.map.gz emd_42484_half_map_2.map.gz emd_42484_half_map_2.map.gz | 59.7 MB 62.1 MB 62.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42484 http://ftp.pdbj.org/pub/emdb/structures/EMD-42484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42484 | HTTPS FTP |
-Validation report
| Summary document |  emd_42484_validation.pdf.gz emd_42484_validation.pdf.gz | 879.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42484_full_validation.pdf.gz emd_42484_full_validation.pdf.gz | 879.2 KB | Display | |
| Data in XML |  emd_42484_validation.xml.gz emd_42484_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_42484_validation.cif.gz emd_42484_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42484 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42484 | HTTPS FTP |
-Related structure data
| Related structure data |  8ur6MC  8um1C  8um2C  8ur3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42484.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42484.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.044 Å | ||||||||||||||||||||||||||||||||||||
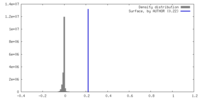
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer map
| File | emd_42484_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
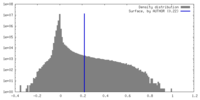
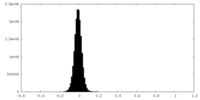
| Density Histograms |
-Half map: #2
| File | emd_42484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
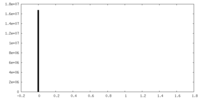
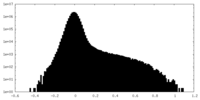
| Density Histograms |
-Half map: #1
| File | emd_42484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
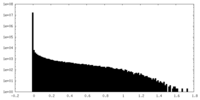
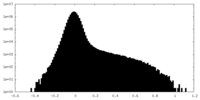
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of OhyA
| Entire | Name: Homodimer of OhyA |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of OhyA
| Supramolecule | Name: Homodimer of OhyA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139.343 KDa |
-Macromolecule #1: Oleate hydratase
| Macromolecule | Name: Oleate hydratase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.892719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MYYSYGNYEA FARPKKPENV ENKSAYLIGS GLASLAAACF LIRDGQMEGS KIHILEELPK AGGSLDGEN MPLKGYVVRG GREMENHFEC LWDLFRSIPS LEIDNASVLD EFYWLNKEDP NYSRCRVIEK QGQRLVTDGD F TLTKTAIK ...String: MGSSHHHHHH SSGLVPRGSH MYYSYGNYEA FARPKKPENV ENKSAYLIGS GLASLAAACF LIRDGQMEGS KIHILEELPK AGGSLDGEN MPLKGYVVRG GREMENHFEC LWDLFRSIPS LEIDNASVLD EFYWLNKEDP NYSRCRVIEK QGQRLVTDGD F TLTKTAIK EILDLCLTNE EDLDDVKITD VFSDDFFNSN FWIYWKTMFA FEPWHSAMEM RRYLMRFVHH ISGLADFSAL KF TKYNQYE SLVLPMVEYL KSHGVQFEYD VKVEDIKIDV TTSQKIAREI LIDRNGNAES IKLTINDLVF VTNGSITESS TYG DNDTPA PPTDELGGSW TLWKNLARQS PEFGNPDKFC QNIPKKSWFV SATSTTNNKE IIDTIESICK RDPLAGKTVT GGII TINDS AWQMSFTINR QQQFKDQPEN EISTWIYALY SDVNGDYIKK PITECSGNEI CQEWLYHLGV STDKIEDLAK HASNT IPVY MPYITSYFMT RAIGDRPLVV PHQSQNLAFI GNFAETERDT VFTTEYSVRT AMEAVYQLLN IDRGIPEVIN SPFDLR VLM DAIYELNDHQ DLREITKDSK MQKLALAGFL KKIKGTYIES LLKEHKLL UniProtKB: Myosin-cross-reactive antigen |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.3 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3248 / Average exposure time: 3.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 79000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model / Details: initial model was a crystal structure |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8ur6: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)