+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | ||||||||||||
 Map data Map data | CryoEM map of S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | immunoglobulin / complex / hemagglutinin / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | ||||||||||||
| Biological species |  Influenza B virus (B/Malaysia/2506/2004) / Influenza B virus (B/Malaysia/2506/2004) /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
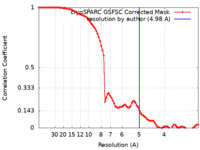
| Method | single particle reconstruction / cryo EM / Resolution: 4.98 Å | ||||||||||||
 Authors Authors | Finney J / Kong S / Walsh Jr RM / Harrison SC / Kelsoe G | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin. Authors: Joel Finney / Annie Park Moseman / Susan Kong / Akiko Watanabe / Shengli Song / Richard M Walsh / Masayuki Kuraoka / Ryutaro Kotaki / E Ashley Moseman / Kevin R McCarthy / Dongmei Liao / ...Authors: Joel Finney / Annie Park Moseman / Susan Kong / Akiko Watanabe / Shengli Song / Richard M Walsh / Masayuki Kuraoka / Ryutaro Kotaki / E Ashley Moseman / Kevin R McCarthy / Dongmei Liao / Xiaoe Liang / Xiaoyan Nie / Olivia Lavidor / Richard Abbott / Stephen C Harrison / Garnett Kelsoe /  Abstract: Phylogenetically and antigenically distinct influenza A and B viruses (IAV and IBV) circulate in human populations, causing widespread morbidity. Antibodies (Abs) that bind epitopes conserved in both ...Phylogenetically and antigenically distinct influenza A and B viruses (IAV and IBV) circulate in human populations, causing widespread morbidity. Antibodies (Abs) that bind epitopes conserved in both IAV and IBV hemagglutinins (HAs) could protect against disease by diverse virus subtypes. Only one reported HA Ab, isolated from a combinatorial display library, protects against both IAV and IBV. Thus, there has been so far no information on the likelihood of finding naturally occurring human Abs that bind HAs of diverse IAV subtypes and IBV lineages. We have now recovered from several unrelated human donors five clonal Abs that bind a conserved epitope preferentially exposed in the postfusion conformation of IAV and IVB HA2. These Abs lack neutralizing activity in vitro but in mice provide strong, IgG subtype-dependent protection against lethal IAV and IBV infections. Strategies to elicit similar Abs routinely might contribute to more effective influenza vaccines. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42149.map.gz emd_42149.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42149-v30.xml emd-42149-v30.xml emd-42149.xml emd-42149.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42149_fsc.xml emd_42149_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42149.png emd_42149.png | 63.5 KB | ||
| Filedesc metadata |  emd-42149.cif.gz emd-42149.cif.gz | 6.3 KB | ||
| Others |  emd_42149_half_map_1.map.gz emd_42149_half_map_1.map.gz emd_42149_half_map_2.map.gz emd_42149_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42149 http://ftp.pdbj.org/pub/emdb/structures/EMD-42149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42149 | HTTPS FTP |
-Related structure data
| Related structure data |  8udgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42149.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42149.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.54688 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: cryoEM Half-map A
| File | emd_42149_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryoEM Half-map B
| File | emd_42149_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of two S1V2-72 Fabs with postfusion HA2
| Entire | Name: Complex of two S1V2-72 Fabs with postfusion HA2 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of two S1V2-72 Fabs with postfusion HA2
| Supramolecule | Name: Complex of two S1V2-72 Fabs with postfusion HA2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#3, #1 Details: Fab fragment generated by proteolytic cleavage of S1V2-72 IgG |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Malaysia/2506/2004) Influenza B virus (B/Malaysia/2506/2004) |
-Macromolecule #1: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Malaysia/2506/2004) Influenza B virus (B/Malaysia/2506/2004) |
| Molecular weight | Theoretical: 17.16916 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GYTSHGAHGV AVAADLKSTQ EAINKITKNL NSLSELEVKN LQRLSGAMDE IHNEILELDE KVDDLRADTI SSQIELAVLL SNEGIINSE DEHLLALERK LKKMLGPSAV DIGNGCFETK HKCNQTCLDR IAAGTFNAGE FSLPTFDSLN ITAASLNDDG UniProtKB: Hemagglutinin |
-Macromolecule #2: S1V2-72 heavy chain
| Macromolecule | Name: S1V2-72 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.135121 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE LKKPGASVKV SCKASGYTFT GNYIHWMRQV PGQGLEWMGW INPRTGDTHH AQKFQGRVDM TRDTSINTAY LELTRLESD DTALYYCARC VFATSQFDPW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: QVQLVQSGAE LKKPGASVKV SCKASGYTFT GNYIHWMRQV PGQGLEWMGW INPRTGDTHH AQKFQGRVDM TRDTSINTAY LELTRLESD DTALYYCARC VFATSQFDPW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDK |
-Macromolecule #3: S1V2-72 light chain
| Macromolecule | Name: S1V2-72 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.755045 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTNSDIG SHNLVSWYQQ HPGKAPKVMI YDDSKRPSGV SNRFSGSKSG STASLTISGL QSEDEADYY CCSYAGSSNW VFGGGTKLTL LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSALTQPASV SGSPGQSITI SCTGTNSDIG SHNLVSWYQQ HPGKAPKVMI YDDSKRPSGV SNRFSGSKSG STASLTISGL QSEDEADYY CCSYAGSSNW VFGGGTKLTL LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 76.12 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8udg: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)