+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mechanically activated ion channel OSCA3.1 in nanodiscs | |||||||||
 Map data Map data | Map postprocessed with LocalDeblur | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mechanically activated ion channel / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast envelope / plasmodesma / plant-type vacuole / mechanosensitive monoatomic ion channel activity / calcium-activated cation channel activity / mRNA binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
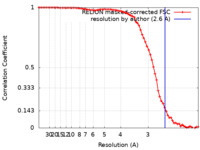
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Jojoa-Cruz S / Lee WH / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli. Authors: Sebastian Jojoa-Cruz / Adrienne E Dubin / Wen-Hsin Lee / Andrew B Ward /  Abstract: The dimeric two-pore OSCA/TMEM63 family has recently been identified as mechanically activated ion channels. Previously, based on the unique features of the structure of OSCA1.2, we postulated the ...The dimeric two-pore OSCA/TMEM63 family has recently been identified as mechanically activated ion channels. Previously, based on the unique features of the structure of OSCA1.2, we postulated the potential involvement of several structural elements in sensing membrane tension (Jojoa-Cruz et al., 2018). Interestingly, while OSCA1, 2, and 3 clades are activated by membrane stretch in cell-attached patches (i.e. they are stretch-activated channels), they differ in their ability to transduce membrane deformation induced by a blunt probe (poking). Here, in an effort to understand the domains contributing to mechanical signal transduction, we used cryo-electron microscopy to solve the structure of (At) OSCA3.1, which, unlike AtOSCA1.2, only produced stretch- but not poke-activated currents in our initial characterization (Murthy et al., 2018). Mutagenesis and electrophysiological assessment of conserved and divergent putative mechanosensitive features of OSCA1.2 reveal a selective disruption of the macroscopic currents elicited by poking without considerable effects on stretch-activated currents (SAC). Our results support the involvement of the amphipathic helix and lipid-interacting residues in the membrane fenestration in the response to poking. Our findings position these two structural elements as potential sources of functional diversity within the family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41911.map.gz emd_41911.map.gz | 22.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41911-v30.xml emd-41911-v30.xml emd-41911.xml emd-41911.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41911_fsc.xml emd_41911_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_41911.png emd_41911.png | 109.4 KB | ||
| Filedesc metadata |  emd-41911.cif.gz emd-41911.cif.gz | 6.9 KB | ||
| Others |  emd_41911_additional_1.map.gz emd_41911_additional_1.map.gz emd_41911_half_map_1.map.gz emd_41911_half_map_1.map.gz emd_41911_half_map_2.map.gz emd_41911_half_map_2.map.gz | 26.4 MB 22.4 MB 22.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41911 http://ftp.pdbj.org/pub/emdb/structures/EMD-41911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41911 | HTTPS FTP |
-Related structure data
| Related structure data |  8u53MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41911.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41911.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map postprocessed with LocalDeblur | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map postprocessed with DeepEMhance
| File | emd_41911_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map postprocessed with DeepEMhance | ||||||||||||
| Projections & Slices |
| ||||||||||||
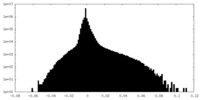
| Density Histograms |
-Half map: Half map 1
| File | emd_41911_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_41911_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OSCA3.1 dimer in nanodisc
| Entire | Name: OSCA3.1 dimer in nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: OSCA3.1 dimer in nanodisc
| Supramolecule | Name: OSCA3.1 dimer in nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 164 KDa |
-Macromolecule #1: CSC1-like protein ERD4
| Macromolecule | Name: CSC1-like protein ERD4 / type: protein_or_peptide / ID: 1 Details: The last 10 residues (GTGTLEVLFQ) are leftover of a linker and protease site. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.878898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGSFLVSL GTSFVIFVIL MLLFTWLSRK SGNAPIYYPN RILKGLEPWE GTSLTRNPFA WMREALTSSE QDVVNLSGVD TAVHFVFLS TVLGIFACSS LLLLPTLLPL AATDNNIKNT KNATDTTSKG TFSQLDNLSM ANITKKSSRL WAFLGAVYWI S LVTYFFLW ...String: MEFGSFLVSL GTSFVIFVIL MLLFTWLSRK SGNAPIYYPN RILKGLEPWE GTSLTRNPFA WMREALTSSE QDVVNLSGVD TAVHFVFLS TVLGIFACSS LLLLPTLLPL AATDNNIKNT KNATDTTSKG TFSQLDNLSM ANITKKSSRL WAFLGAVYWI S LVTYFFLW KAYKHVSSLR AQALMSADVK PEQFAILVRD MPAPPDGQTQ KEFIDSYFRE IYPETFYRSL VATEN(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)K KKLARAEAIL AATNNRPTN KTGFCGLVGK QVD(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)AE KQQTAAVVFF TTRVAAASAA QSL HCQMVD KWTVTEAPEP RQLLWQNLNI KLFSRIIRQY FIYFFVAVTI LFYMIPIAFV SAITTLKNLQ RIIPFIKPVV EITA IRTVL ESFLPQIALI VFLAMLPKLL LFLSKAEGIP SQSHAIRAAS GKYFYFSVFN VFIGVTLAGT LFNTVKDIAK NPKLD MIIN LLATSLPKSA TFFLTYVALK FFIGYGLELS RIIPLIIFHL KKKYLCKTEA EVKEAWYPGD LSYATRVPGD MLILTI TFC YSVIAPLILI FGITYFGLGW LVLRNQALKV YVPSYESYGR MWPHIHQRIL AALFLFQVVM FGYLGAKTFF YTALVIP LI ITSLIFGYVC RQKFYGGFEH TALEVACREL KQSPDLEEIF RAYIPHSLSS HKPEEHEFKG AMSRYQDFNA IAGVGTGT L EVLFQ UniProtKB: Hyperosmolality-gated Ca2+ permeable channel 3.1 |
-Macromolecule #2: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 2 / Formula: CPL |
|---|---|
| Molecular weight | Theoretical: 758.06 Da |
| Chemical component information |  ChemComp-CPL: |
-Macromolecule #3: [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl...
| Macromolecule | Name: [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate type: ligand / ID: 3 / Number of copies: 2 / Formula: 82T |
|---|---|
| Molecular weight | Theoretical: 481.603 Da |
| Chemical component information |  ChemComp-82T: |
-Macromolecule #4: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 4 / Number of copies: 20 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 1-38 / Number grids imaged: 1 / Number real images: 8375 / Average electron dose: 50.0 e/Å2 Details: Only micrographs with a CTF estimate of 2.6A or better were used for processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.4 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
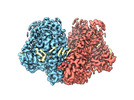
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)