+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DdmD dimer in complex with ssDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DdmD / helicase / nuclease / IMMUNE SYSTEM / IMMUNE SYSTEM-DNA complex | |||||||||
| Function / homology | Helicase/UvrB N-terminal domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Bravo JPK / Taylor DW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Plasmid targeting and destruction by the DdmDE bacterial defence system. Authors: Jack P K Bravo / Delisa A Ramos / Rodrigo Fregoso Ocampo / Caiden Ingram / David W Taylor /   Abstract: Although eukaryotic Argonautes have a pivotal role in post-transcriptional gene regulation through nucleic acid cleavage, some short prokaryotic Argonaute variants (pAgos) rely on auxiliary nuclease ...Although eukaryotic Argonautes have a pivotal role in post-transcriptional gene regulation through nucleic acid cleavage, some short prokaryotic Argonaute variants (pAgos) rely on auxiliary nuclease factors for efficient foreign DNA degradation. Here we reveal the activation pathway of the DNA defence module DdmDE system, which rapidly eliminates small, multicopy plasmids from the Vibrio cholerae seventh pandemic strain (7PET). Through a combination of cryo-electron microscopy, biochemistry and in vivo plasmid clearance assays, we demonstrate that DdmE is a catalytically inactive, DNA-guided, DNA-targeting pAgo with a distinctive insertion domain. We observe that the helicase-nuclease DdmD transitions from an autoinhibited, dimeric complex to a monomeric state upon loading of single-stranded DNA targets. Furthermore, the complete structure of the DdmDE-guide-target handover complex provides a comprehensive view into how DNA recognition triggers processive plasmid destruction. Our work establishes a mechanistic foundation for how pAgos utilize ancillary factors to achieve plasmid clearance, and provides insights into anti-plasmid immunity in bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41785.map.gz emd_41785.map.gz | 255.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41785-v30.xml emd-41785-v30.xml emd-41785.xml emd-41785.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41785.png emd_41785.png | 77.9 KB | ||
| Filedesc metadata |  emd-41785.cif.gz emd-41785.cif.gz | 6.7 KB | ||
| Others |  emd_41785_half_map_1.map.gz emd_41785_half_map_1.map.gz emd_41785_half_map_2.map.gz emd_41785_half_map_2.map.gz | 474.4 MB 474.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41785 http://ftp.pdbj.org/pub/emdb/structures/EMD-41785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41785 | HTTPS FTP |
-Related structure data
| Related structure data |  8u0uMC  8u0jC  8u0wC  8u3kC  9bqvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41785.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41785.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41785_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
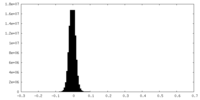
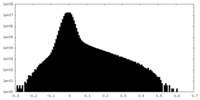
| Density Histograms |
-Half map: #2
| File | emd_41785_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
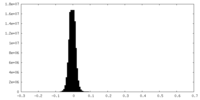
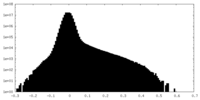
| Density Histograms |
- Sample components
Sample components
-Entire : DdmD
| Entire | Name: DdmD |
|---|---|
| Components |
|
-Supramolecule #1: DdmD
| Supramolecule | Name: DdmD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Helicase/UvrB N-terminal domain-containing protein
| Supramolecule | Name: Helicase/UvrB N-terminal domain-containing protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Helicase/UvrB N-terminal domain-containing protein
| Macromolecule | Name: Helicase/UvrB N-terminal domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 136.145297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNVSIEEFTH FDFQLVPEPS PLDLVITESL KNHIEVNGVK SGALLPLPFQ TGIGKTYTAL NFLLQQMLEQ VRSELKEENT GKKSKRLLY YVTDSVDNVV SAKADLLKLI EKQTVKGEPR FTLEQQEYLK AQIVHLPNQS EQLLQCSDAV LNDVLIGFNL N AERDVQAE ...String: MNVSIEEFTH FDFQLVPEPS PLDLVITESL KNHIEVNGVK SGALLPLPFQ TGIGKTYTAL NFLLQQMLEQ VRSELKEENT GKKSKRLLY YVTDSVDNVV SAKADLLKLI EKQTVKGEPR FTLEQQEYLK AQIVHLPNQS EQLLQCSDAV LNDVLIGFNL N AERDVQAE WSAISGLRRH ASNPEVKISL NRQAGYFYRN LIDRLQKKQK GADRVLLSGS LLASVETLLP GEKIRNGSAH VA FLTTSKF LKGFHNTRSR YSPLRDLSGA VLIIDEIDKQ NQVILSELCK QQAQDLIWAI RTLRANFRDH QLESSPRYDK IED LFEPLR ERLEEFGTNW NLAFAFNTEG ANLNERPVRL FSDRSFTHVS SATHKLSLKS DFLRRKNLIF SDEKVEGSLI EKHG LLTRF VNEADVIYQW FLGTMRKAVF QYWENVRGLE IEVRENRSLE GTFQEAVQSL LTHFNLQEFE SAVYESFDTR GLRQS AGGK ANKLSSSKSY HHTGLKLVEV AHNQGTRDTV NCKASFLNTS PSGVLADMVD AGAVILGISA TARADTVIHN FDFKYL NER LGNKLLSLSR EQKQRVNNYY HSRRNYKDNG VVLTVKYLNS RDAFLDALLE EYKPEARSSH FILNHYLGIA ESEQAFV RS WLSKLLASIK AFISSPDNRY MLSLLNRTLD TTRQNINDFI QFCCDKWAKE FNVKTKTFFG VNADWMRLVG YDEISKHL N TELGKVVVFS TYASMGAGKN PDYAVNLALE GESLISVADV TYSTQLRSDI DSIYLEKPTQ LLLSDDYSHT ANQLCQFHQ ILSLQENGEL SPKSAENWCR QQLMGMSRER SLQQYHQTSD YQSAVRKYIE QAVGRAGRTS LKRKQILLFV DSGLKEILAE ESRDPSLFS HEYVALVNKA KSAGKSIVED RAVRRLFNLA QRNNKDGMLS IKALVHRLHN QPASKSDIQE WQDIRTQLLR Y PTVAFQPE RFNRLYLQSM TKGYYRYQGN LDGDPNSFEF FDRVPYGDMV SEEDCSLATL VQNQYVRPWF ERKGFACSWQ KE ANVMTPI MFTNIYKGAL GEQAVEAVLT AFDFTFEEVP NSIYERFDNR VIFAGIEQPI WLDSKYWKHE GNESSEGYSS KIA LVEEEF GPSKFIYVNA LGDTSKPIRY LNSCFVETSP QLAKVIEIPA LIDDSNADTN RTAVQELIKW LHHS UniProtKB: Helicase/UvrB N-terminal domain-containing protein |
-Macromolecule #2: DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*T)-3') / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.310177 KDa |
| Sequence | String: (DT)(DT)(DA)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)