+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | |||||||||
 Map data Map data | Sharpened map resulting from Non-Uniform refinement (C2) in cryoSPARC. This is the primary map used for model building and PDB validation. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Heparan-alpha-glucosaminide N-acetyltransferase / Transmembrane protein 76 / membrane protein / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationheparan-alpha-glucosaminide N-acetyltransferase / heparan-alpha-glucosaminide N-acetyltransferase activity / MPS IIIC - Sanfilippo syndrome C / heparan sulfate proteoglycan catabolic process / HS-GAG degradation / lysosomal transport / acyltransferase activity / protein complex oligomerization / tertiary granule membrane / specific granule membrane ...heparan-alpha-glucosaminide N-acetyltransferase / heparan-alpha-glucosaminide N-acetyltransferase activity / MPS IIIC - Sanfilippo syndrome C / heparan sulfate proteoglycan catabolic process / HS-GAG degradation / lysosomal transport / acyltransferase activity / protein complex oligomerization / tertiary granule membrane / specific granule membrane / lysosomal lumen / bioluminescence / generation of precursor metabolites and energy / lysosomal membrane / Neutrophil degranulation / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
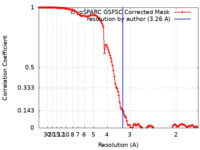
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Navratna V / Kumar A / Mosalaganti S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2024 Title: Structure of the human heparan-α-glucosaminide -acetyltransferase (HGSNAT). Authors: Vikas Navratna / Arvind Kumar / Jaimin K Rana / Shyamal Mosalaganti /  Abstract: Degradation of heparan sulfate (HS), a glycosaminoglycan (GAG) comprised of repeating units of -acetylglucosamine and glucuronic acid, begins in the cytosol and is completed in the lysosomes. ...Degradation of heparan sulfate (HS), a glycosaminoglycan (GAG) comprised of repeating units of -acetylglucosamine and glucuronic acid, begins in the cytosol and is completed in the lysosomes. Acetylation of the terminal non-reducing amino group of a-D-glucosamine of HS is essential for its complete breakdown into monosaccharides and free sulfate. Heparan-a-glucosaminide -acetyltransferase (HGSNAT), a resident of the lysosomal membrane, catalyzes this essential acetylation reaction by accepting and transferring the acetyl group from cytosolic acetyl-CoA to terminal a-D-glucosamine of HS in the lysosomal lumen. Mutation-induced dysfunction in HGSNAT causes abnormal accumulation of HS within the lysosomes and leads to an autosomal recessive neurodegenerative lysosomal storage disorder called mucopolysaccharidosis IIIC (MPS IIIC). There are no approved drugs or treatment strategies to cure or manage the symptoms of, MPS IIIC. Here, we use cryo-electron microscopy (cryo-EM) to determine a high-resolution structure of the HGSNAT-acetyl-CoA complex, the first step in HGSNAT catalyzed acetyltransferase reaction. In addition, we map the known MPS IIIC mutations onto the structure and elucidate the molecular basis for mutation-induced HGSNAT dysfunction. #1:  Journal: eLife / Year: 2024 Journal: eLife / Year: 2024Title: Structure of the human heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT) Authors: Navratna V / Kumar A / Mosalaganti S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41620.map.gz emd_41620.map.gz | 167.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41620-v30.xml emd-41620-v30.xml emd-41620.xml emd-41620.xml | 44 KB 44 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41620_fsc.xml emd_41620_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41620.png emd_41620.png | 89.4 KB | ||
| Filedesc metadata |  emd-41620.cif.gz emd-41620.cif.gz | 7.9 KB | ||
| Others |  emd_41620_additional_1.map.gz emd_41620_additional_1.map.gz emd_41620_additional_10.map.gz emd_41620_additional_10.map.gz emd_41620_additional_11.map.gz emd_41620_additional_11.map.gz emd_41620_additional_2.map.gz emd_41620_additional_2.map.gz emd_41620_additional_3.map.gz emd_41620_additional_3.map.gz emd_41620_additional_4.map.gz emd_41620_additional_4.map.gz emd_41620_additional_5.map.gz emd_41620_additional_5.map.gz emd_41620_additional_6.map.gz emd_41620_additional_6.map.gz emd_41620_additional_7.map.gz emd_41620_additional_7.map.gz emd_41620_additional_8.map.gz emd_41620_additional_8.map.gz emd_41620_additional_9.map.gz emd_41620_additional_9.map.gz emd_41620_half_map_1.map.gz emd_41620_half_map_1.map.gz emd_41620_half_map_2.map.gz emd_41620_half_map_2.map.gz | 786.6 KB 791.2 KB 779.1 KB 779.1 KB 779.1 KB 167.7 MB 164.8 MB 779.1 KB 164.8 MB 88.5 MB 165.2 MB 164.8 MB 164.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41620 http://ftp.pdbj.org/pub/emdb/structures/EMD-41620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41620 | HTTPS FTP |
-Related structure data
| Related structure data |  8tu9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41620.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41620.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map resulting from Non-Uniform refinement (C2) in cryoSPARC. This is the primary map used for model building and PDB validation. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
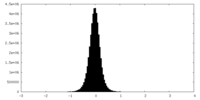
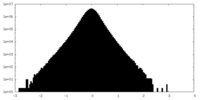
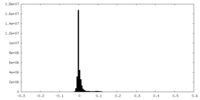
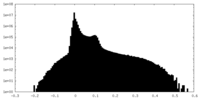
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: Mask (auto-tightening) used for Non-Uniform refinement in cryoSPARC.
+Additional map: Half map (half-map A) resulting from local refinement...
+Additional map: Half map (half-map B) resulting from local refinement...
+Additional map: Mask used for Non-uniform refinement (C2) in cryoSPARC.
+Additional map: Mask used for Non-Uniform refinement (C2) in cryoSPARC.
+Additional map: Mask Used for refinement (C1) during local refinement...
+Additional map: Unsharpened map resulting from Non-Uniform refinement (C2) in...
+Additional map: Composite map for model validation generated from focused...
+Additional map: Mask (auto-tightening) used for refinement during local refinement...
+Additional map: Mask used for refinement during local refinement (C1)...
+Additional map: Unsharpened map resulting from local refinement (C1) in...
+Half map: Half map (half-map A) resulting from Non-Uniform refinement...
+Half map: Half map (half-map B) resulting from Non-Uniform refinement...
- Sample components
Sample components
-Entire : Heparan acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT)
| Entire | Name: Heparan acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT) |
|---|---|
| Components |
|
-Supramolecule #1: Heparan acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT)
| Supramolecule | Name: Heparan acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Full length (Isoform 2) human HGSNAT expressed as a recombinant fusion protein with N-terminal GFP, in mammalian cells. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Lysosomes / Location in cell: Lysosomal membrane Homo sapiens (human) / Organelle: Lysosomes / Location in cell: Lysosomal membrane |
| Molecular weight | Theoretical: 100.7 KDa |
-Macromolecule #1: Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-...
| Macromolecule | Name: Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: heparan-alpha-glucosaminide N-acetyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.817172 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MWSHPQFEKG SGVSKGEELF TGVVPILVEL DGDVNGHKFS VSGEGEGDAT YGKLTLKFIC TTGKLPVPWP TLVTTLTYGV QCFSRYPDH MKQHDFFKSA MPEGYVQERT IFFKDDGNYK TRAEVKFEGD TLVNRIELKG IDFKEDGNIL GHKLEYNYNS H NVYIMADK ...String: MWSHPQFEKG SGVSKGEELF TGVVPILVEL DGDVNGHKFS VSGEGEGDAT YGKLTLKFIC TTGKLPVPWP TLVTTLTYGV QCFSRYPDH MKQHDFFKSA MPEGYVQERT IFFKDDGNYK TRAEVKFEGD TLVNRIELKG IDFKEDGNIL GHKLEYNYNS H NVYIMADK QKNGIKVNFK IRHNIEDGSV QLADHYQQNT PIGDGPVLLP DNHYLSTQSK LSKDPNEKRD HMVLLEFVTA AG ITLGMDE LYKSGLRSGL EVLFQGPEFE FMSGAGRALA ALLLAASVLS AALLAPGGSS GRDAQAAPPR DLDKKRHAEL KMD QALLLI HNELLWTNLT VYWKSECCYH CLFQVLVNVP QSPKAGKPSA AAASVSTQHG SILQLNDTLE EKEVCRLEYR FGEF GNYSL LVKNIHNGVS EIACDLAVNE DPVDSNLPVS IAFLIGLAVI IVISFLRLLL SLDDFNNWIS KAISSRETDR LINSE LGSP SRTDPLDGDV QPATWRLSAL PPRLRSVDTF RGIALILMVF VNYGGGKYWY FKHASWNGLT VADLVFPWFV FIMGSS IFL SMTSILQRGC SKFRLLGKIA WRSFLLICIG IIIVNPNYCL GPLSWDKVRI PGVLQRLGVT YFVVAVLELL FAKPVPE HC ASERSCLSLR DITSSWPQWL LILVLEGLWL GLTFLLPVPG CPTGYLGPGG IGDFGKYPNC TGGAAGYIDR LLLGDDHL Y QHPSSAVLYH TEVAYDPEGI LGTINSIVMA FLGVQAGKIL LYYKARTKDI LIRFTAWCCI LGLISVALTK VSENEGFIP VNKNLWSLSY VTTLSSFAFF ILLVLYPVVD VKGLWTGTPF FYPGMNSILV YVGHEVFENY FPFQWKLKDN QSHKEHLTQN IVATALWVL IAYILYRKKI FWKI UniProtKB: Green fluorescent protein, Heparan-alpha-glucosaminide N-acetyltransferase |
-Macromolecule #2: ACETYL COENZYME *A
| Macromolecule | Name: ACETYL COENZYME *A / type: ligand / ID: 2 / Number of copies: 2 / Formula: ACO |
|---|---|
| Molecular weight | Theoretical: 809.571 Da |
| Chemical component information |  ChemComp-ACO: |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Pressure: 0.026000000000000002 kPa / Details: 15 mA current | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | HGSNAT in digitonin micelle, purified by Strep-Tactin affinity chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 10000 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: Made using Model angelo |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-8tu9: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)