+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of GPR61- | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / inverse agonist / membrane protein / BRIL / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-independent adenylate cyclase-activating G protein-coupled receptor signaling pathway / arrestin family protein binding / electron transport chain / G protein-coupled receptor activity / electron transfer activity / periplasmic space / receptor complex / endosome / endosome membrane / iron ion binding ...ligand-independent adenylate cyclase-activating G protein-coupled receptor signaling pathway / arrestin family protein binding / electron transport chain / G protein-coupled receptor activity / electron transfer activity / periplasmic space / receptor complex / endosome / endosome membrane / iron ion binding / G protein-coupled receptor signaling pathway / heme binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
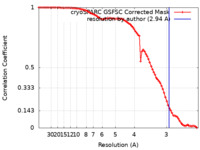
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Lees JA / Dias JM / Han S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism. Authors: Joshua A Lees / João M Dias / Francis Rajamohan / Jean-Philippe Fortin / Rebecca O'Connor / Jimmy X Kong / Emily A G Hughes / Ethan L Fisher / Jamison B Tuttle / Gabrielle Lovett / Bethany ...Authors: Joshua A Lees / João M Dias / Francis Rajamohan / Jean-Philippe Fortin / Rebecca O'Connor / Jimmy X Kong / Emily A G Hughes / Ethan L Fisher / Jamison B Tuttle / Gabrielle Lovett / Bethany L Kormos / Rayomand J Unwalla / Lei Zhang / Anne-Marie Dechert Schmitt / Dahui Zhou / Michael Moran / Kimberly A Stevens / Kimberly F Fennell / Alison E Varghese / Andrew Maxwell / Emmaline E Cote / Yuan Zhang / Seungil Han /  Abstract: GPR61 is an orphan GPCR related to biogenic amine receptors. Its association with phenotypes relating to appetite makes it of interest as a druggable target to treat disorders of metabolism and body ...GPR61 is an orphan GPCR related to biogenic amine receptors. Its association with phenotypes relating to appetite makes it of interest as a druggable target to treat disorders of metabolism and body weight, such as obesity and cachexia. To date, the lack of structural information or a known biological ligand or tool compound has hindered comprehensive efforts to study GPR61 structure and function. Here, we report a structural characterization of GPR61, in both its active-like complex with heterotrimeric G protein and in its inactive state. Moreover, we report the discovery of a potent and selective small-molecule inverse agonist against GPR61 and structural elucidation of its allosteric binding site and mode of action. These findings offer mechanistic insights into an orphan GPCR while providing both a structural framework and tool compound to support further studies of GPR61 function and modulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41145.map.gz emd_41145.map.gz | 56.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41145-v30.xml emd-41145-v30.xml emd-41145.xml emd-41145.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41145_fsc.xml emd_41145_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_41145.png emd_41145.png | 96.2 KB | ||
| Filedesc metadata |  emd-41145.cif.gz emd-41145.cif.gz | 7.1 KB | ||
| Others |  emd_41145_half_map_1.map.gz emd_41145_half_map_1.map.gz emd_41145_half_map_2.map.gz emd_41145_half_map_2.map.gz | 55.4 MB 55.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41145 http://ftp.pdbj.org/pub/emdb/structures/EMD-41145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41145 | HTTPS FTP |
-Related structure data
| Related structure data |  8tb7MC  8tb0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41145.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41145.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.18 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
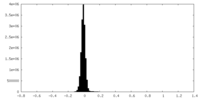
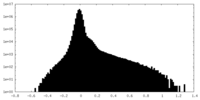
| Density Histograms |
-Half map: #1
| File | emd_41145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
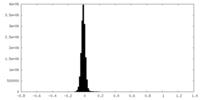
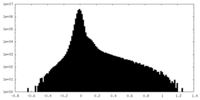
| Density Histograms |
- Sample components
Sample components
-Entire : GPR61-ICL3-BRIL chimera in complex with anti-BRIL Fab24 BAK5 and ...
| Entire | Name: GPR61-ICL3-BRIL chimera in complex with anti-BRIL Fab24 BAK5 and hinge-binding nanobody bound to inverse agonist Compound 1 |
|---|---|
| Components |
|
-Supramolecule #1: GPR61-ICL3-BRIL chimera in complex with anti-BRIL Fab24 BAK5 and ...
| Supramolecule | Name: GPR61-ICL3-BRIL chimera in complex with anti-BRIL Fab24 BAK5 and hinge-binding nanobody bound to inverse agonist Compound 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: G-protein coupled receptor 61,Soluble cytochrome b562
| Macromolecule | Name: G-protein coupled receptor 61,Soluble cytochrome b562 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.991902 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYPYDVPDYA ESSPIPQSSG NSSTLGRVPQ TPGPSTASGV PEVGLRDVAS ESVALFFMLL LDLTAVAGNA AVMAVIAKTP ALRKFVFVF HLCLVDLLAA LTLMPLAMLS SSALFDHALF GEVACRLYLF LSVCFVSLAI LSVSAINVER YYYVVHPMRY E VRMTLGLV ...String: MYPYDVPDYA ESSPIPQSSG NSSTLGRVPQ TPGPSTASGV PEVGLRDVAS ESVALFFMLL LDLTAVAGNA AVMAVIAKTP ALRKFVFVF HLCLVDLLAA LTLMPLAMLS SSALFDHALF GEVACRLYLF LSVCFVSLAI LSVSAINVER YYYVVHPMRY E VRMTLGLV ASVLVGVWVK ALAMASVPVL GRVSWEEGAP SVPPGCSLQW SHSAYCQLFV VVFAVLYFLL PLLLILVVYC SM FRARRQL ADLEDNWETL NDNLKVIEKA DNAAQVKDAL TKMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QID DALKLA NEGKVKEAQA AAEQLKTTRN AYIQKYLERA RSTLQKEVKA AVVLLAVGGQ FLLCWLPYFS FHLYVALSAQ PIST GQVES VVTWIGYFCF TSNPFFYGCL NRQIRGELSK QFVCFFKPAP EEELRLPSRE GSIEENFLQF LQGTGCPSES WVSRP LPSP KQEPPAVDFR IPGQIAEETS EFLEQQLTSD IIMSDSYLRP AASPRLESGS DYKDDDDAK UniProtKB: G-protein coupled receptor 61, Soluble cytochrome b562, G-protein coupled receptor 61 |
-Macromolecule #2: Fab24 BAK5 heavy chain
| Macromolecule | Name: Fab24 BAK5 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.888203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQYLYYS LVTFGQGTKV EIKRTVAAPS VFIFPPSDSQ LKSGTASVVC L LNNFYPRE ...String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQYLYYS LVTFGQGTKV EIKRTVAAPS VFIFPPSDSQ LKSGTASVVC L LNNFYPRE AKVQWKVDNA LQSGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE MK KNIAFLL ASMFVFSIAT NAYAEISEVQ LVESGGGLVQ PGGSLRLSCA ASGFNVRSFS IHWVRQAPGK GLEWVAYISS SSG STSYAD SVKGRFTISA DTSKNTAYLQ MNSLRAEDTA VYYCARWGYW PGEPWWKAFD YWGQGTLVTV SSASTKGPSV FPLA PSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNT KVDK KVEPKS |
-Macromolecule #3: Fab hinge-binding nanobody
| Macromolecule | Name: Fab hinge-binding nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.276541 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSQVQLQESG GGLVQPGGSL RLSCAASGRT ISRYAMSWFR QAPGKEREFV AVARRSGDGA FYADSVQGRF TVSRDDAKTV YLQMNSLKP EDTAVYYCAI DSDTFYSGSY DYWGQGTQVT VSS |
-Macromolecule #4: Fab24 BAK5 light chain
| Macromolecule | Name: Fab24 BAK5 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.398066 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: MDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRG |
-Macromolecule #5: 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylp...
| Macromolecule | Name: 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide type: ligand / ID: 5 / Number of copies: 1 / Formula: ZOB |
|---|---|
| Molecular weight | Theoretical: 524.541 Da |
| Chemical component information | 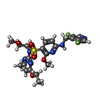 ChemComp-ZOB: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)