[English] 日本語
 Yorodumi
Yorodumi- EMDB-41126: Cryo-EM structure of the human CLC-2 chloride channel transmembra... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Chloride / Channel / Inhibitor / Protein / Voltage gated / TRANSPORT PROTEIN-INHIBITOR complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of aldosterone biosynthetic process / cell differentiation involved in salivary gland development / astrocyte end-foot / acinar cell differentiation / voltage-gated chloride channel activity / dendritic spine membrane / chloride transport / phagocytosis, engulfment / positive regulation of oligodendrocyte differentiation / chloride channel complex ...regulation of aldosterone biosynthetic process / cell differentiation involved in salivary gland development / astrocyte end-foot / acinar cell differentiation / voltage-gated chloride channel activity / dendritic spine membrane / chloride transport / phagocytosis, engulfment / positive regulation of oligodendrocyte differentiation / chloride channel complex / lung development / Stimuli-sensing channels / myelin sheath / retina development in camera-type eye / basolateral plasma membrane / perikaryon / postsynaptic membrane / axon / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||||||||
 Authors Authors | Xu M / Neelands T / Powers AS / Liu Y / Miller S / Pintilie G / Du Bois J / Dror RO / Chiu W / Maduke M | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism. Authors: Mengyuan Xu / Torben Neelands / Alexander S Powers / Yan Liu / Steven D Miller / Grigore D Pintilie / J Du Bois / Ron O Dror / Wah Chiu / Merritt Maduke /  Abstract: CLC-2 is a voltage-gated chloride channel that contributes to electrical excitability and ion homeostasis in many different tissues. Among the nine mammalian CLC homologs, CLC-2 is uniquely activated ...CLC-2 is a voltage-gated chloride channel that contributes to electrical excitability and ion homeostasis in many different tissues. Among the nine mammalian CLC homologs, CLC-2 is uniquely activated by hyperpolarization, rather than depolarization, of the plasma membrane. The molecular basis for the divergence in polarity of voltage gating among closely related homologs has been a long-standing mystery, in part because few CLC channel structures are available. Here, we report cryoEM structures of human CLC-2 at 2.46 - 2.76 Å, in the presence and absence of the selective inhibitor AK-42. AK-42 binds within the extracellular entryway of the Cl-permeation pathway, occupying a pocket previously proposed through computational docking studies. In the apo structure, we observed two distinct conformations involving rotation of one of the cytoplasmic C-terminal domains (CTDs). In the absence of CTD rotation, an intracellular N-terminal 15-residue hairpin peptide nestles against the TM domain to physically occlude the Cl-permeation pathway. This peptide is highly conserved among species variants of CLC-2 but is not present in other CLC homologs. Previous studies suggested that the N-terminal domain of CLC-2 influences channel properties via a "ball-and-chain" gating mechanism, but conflicting data cast doubt on such a mechanism, and thus the structure of the N-terminal domain and its interaction with the channel has been uncertain. Through electrophysiological studies of an N-terminal deletion mutant lacking the 15-residue hairpin peptide, we support a model in which the N-terminal hairpin of CLC-2 stabilizes a closed state of the channel by blocking the cytoplasmic Cl-permeation pathway. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41126.map.gz emd_41126.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41126-v30.xml emd-41126-v30.xml emd-41126.xml emd-41126.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41126.png emd_41126.png | 129.4 KB | ||
| Filedesc metadata |  emd-41126.cif.gz emd-41126.cif.gz | 6.3 KB | ||
| Others |  emd_41126_half_map_1.map.gz emd_41126_half_map_1.map.gz emd_41126_half_map_2.map.gz emd_41126_half_map_2.map.gz | 59 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41126 http://ftp.pdbj.org/pub/emdb/structures/EMD-41126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41126 | HTTPS FTP |
-Validation report
| Summary document |  emd_41126_validation.pdf.gz emd_41126_validation.pdf.gz | 761.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41126_full_validation.pdf.gz emd_41126_full_validation.pdf.gz | 761.3 KB | Display | |
| Data in XML |  emd_41126_validation.xml.gz emd_41126_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_41126_validation.cif.gz emd_41126_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41126 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41126 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41126 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41126 | HTTPS FTP |
-Related structure data
| Related structure data |  8ta2MC  8ta3C  8ta4C  8ta5C  8ta6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41126.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41126.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.946 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41126_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
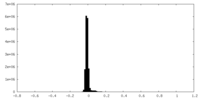
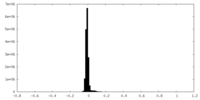
| Density Histograms |
-Half map: #1
| File | emd_41126_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
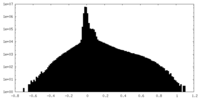
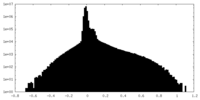
| Density Histograms |
- Sample components
Sample components
-Entire : Chloride channel protein 2 with inhibitor AK-42
| Entire | Name: Chloride channel protein 2 with inhibitor AK-42 |
|---|---|
| Components |
|
-Supramolecule #1: Chloride channel protein 2 with inhibitor AK-42
| Supramolecule | Name: Chloride channel protein 2 with inhibitor AK-42 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Chloride channel protein 2
| Macromolecule | Name: Chloride channel protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.390672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GEDWIFLVLL GLLMALVSWV MDYAIAACLQ AQQWMSRGLN TSILLQYLAW VTYPVVLITF SAGFTQILAP QAVGSGIPEM KTILRGVVL KEYLTLKTFI AKVIGLTCAL GSGMPLGKEG PFVHIASMCA ALLSKFLSLF GGIYENESRN TEMLAAACAV G VGCCFAAP ...String: GEDWIFLVLL GLLMALVSWV MDYAIAACLQ AQQWMSRGLN TSILLQYLAW VTYPVVLITF SAGFTQILAP QAVGSGIPEM KTILRGVVL KEYLTLKTFI AKVIGLTCAL GSGMPLGKEG PFVHIASMCA ALLSKFLSLF GGIYENESRN TEMLAAACAV G VGCCFAAP IGGVLFSIEV TSTFFAVRNY WRGFFAATFS AFIFRVLAVW NRDEETITAL FKTRFRLDFP FDLQELPAFA VI GIASGFG GALFVYLNRK IVQVMRKQKT INRFLMRKRL LFPALVTLLI STLTFPPGFG QFMAGQLSQK ETLVTLFDNR TWV RQGLVE ELEPPSTSQA WNPPRANVFL TLVIFILMKF WMSALATTIP VPCGAFMPVF VIGAAFGRLV GESMAAWFPD GIHT DSSTY RIVPGGYAVV GAAALAGAVT HTVSTAVIVF ELTGQIAHIL PVMIAVILAN AVAQSLQPSL YDSIIRIKKL PYLP UniProtKB: Chloride channel protein 2 |
-Macromolecule #2: 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-ca...
| Macromolecule | Name: 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: GH6 |
|---|---|
| Molecular weight | Theoretical: 389.232 Da |
| Chemical component information | 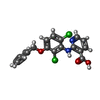 ChemComp-GH6: |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 14300 / Average exposure time: 5.6 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)