[English] 日本語
 Yorodumi
Yorodumi- EMDB-40719: particulate methane monooxygeanse treated with potassium cyanide ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | particulate methane monooxygeanse treated with potassium cyanide and copper reloaded | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Metalloenzyme / Membrane Protein / Nanodiscs / Oxidoreductase | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmethane monooxygenase (particulate) / methane monooxygenase (soluble) / methane monooxygenase [NAD(P)H] activity / monooxygenase activity / metal ion binding / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Methylococcus capsulatus (bacteria) Methylococcus capsulatus (bacteria) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||||||||||||||
 Authors Authors | Tucci FJ / Jodts RJ / Rosenzweig AC | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Catal / Year: 2023 Journal: Nat Catal / Year: 2023Title: Product analog binding identifies the copper active site of particulate methane monooxygenase. Authors: Frank J Tucci / Richard J Jodts / Brian M Hoffman / Amy C Rosenzweig /  Abstract: Nature's primary methane-oxidizing enzyme, the membrane-bound particulate methane monooxygenase (pMMO), catalyzes the oxidation of methane to methanol. pMMO activity requires copper, and decades of ...Nature's primary methane-oxidizing enzyme, the membrane-bound particulate methane monooxygenase (pMMO), catalyzes the oxidation of methane to methanol. pMMO activity requires copper, and decades of structural and spectroscopic studies have sought to identify the active site among three candidates: the Cu, Cu, and Cu sites. Challenges associated with the isolation of active pMMO have hindered progress toward locating its catalytic center. However, reconstituting pMMO into native lipid nanodiscs stabilizes its structure and recovers its activity. Here, these active samples were incubated with 2,2,2,-trifluoroethanol (TFE), a product analog that serves as a readily visualized active-site probe. Interactions of TFE with the Cu site were observed by both pulsed ENDOR spectroscopy and cryoEM, implicating Cu and the surrounding hydrophobic pocket as the likely site of methane oxidation. Use of these orthogonal techniques on parallel samples is a powerful approach that can circumvent difficulties in interpreting metalloenzyme cryoEM maps. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40719.map.gz emd_40719.map.gz | 34.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40719-v30.xml emd-40719-v30.xml emd-40719.xml emd-40719.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40719.png emd_40719.png | 115.6 KB | ||
| Filedesc metadata |  emd-40719.cif.gz emd-40719.cif.gz | 5.8 KB | ||
| Others |  emd_40719_half_map_1.map.gz emd_40719_half_map_1.map.gz emd_40719_half_map_2.map.gz emd_40719_half_map_2.map.gz | 475.1 MB 475.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40719 http://ftp.pdbj.org/pub/emdb/structures/EMD-40719 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40719 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40719 | HTTPS FTP |
-Validation report
| Summary document |  emd_40719_validation.pdf.gz emd_40719_validation.pdf.gz | 757.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40719_full_validation.pdf.gz emd_40719_full_validation.pdf.gz | 757.3 KB | Display | |
| Data in XML |  emd_40719_validation.xml.gz emd_40719_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_40719_validation.cif.gz emd_40719_validation.cif.gz | 22.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40719 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40719 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40719 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40719 | HTTPS FTP |
-Related structure data
| Related structure data |  8sr4MC  8oyiC  8sqwC  8sr1C  8sr2C  8sr5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40719.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40719.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.5291 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40719_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
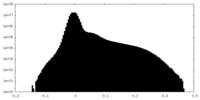
| Projections & Slices |
| ||||||||||||
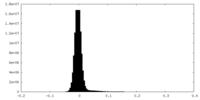
| Density Histograms |
-Half map: #1
| File | emd_40719_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
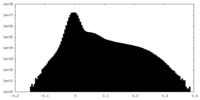
| Projections & Slices |
| ||||||||||||
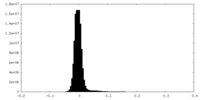
| Density Histograms |
- Sample components
Sample components
-Entire : particulate methane monooxygenase
| Entire | Name: particulate methane monooxygenase |
|---|---|
| Components |
|
-Supramolecule #1: particulate methane monooxygenase
| Supramolecule | Name: particulate methane monooxygenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Methylococcus capsulatus (bacteria) / Strain: Bath Methylococcus capsulatus (bacteria) / Strain: Bath |
| Molecular weight | Theoretical: 337.11 kDa/nm |
-Macromolecule #1: Particulate methane monooxygenase alpha subunit
| Macromolecule | Name: Particulate methane monooxygenase alpha subunit / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methylococcus capsulatus (bacteria) / Strain: Bath Methylococcus capsulatus (bacteria) / Strain: Bath |
| Molecular weight | Theoretical: 42.832887 KDa |
| Sequence | String: HGEKSQAAFM RMRTIHWYDL SWSKEKVKIN ETVEIKGKFH VFEGWPETVD EPDVAFLNVG MPGPVFIRKE SYIGGQLVPR SVRLEIGKT YDFRVVLKAR RPGDWHVHTM MNVQGGGPII GPGKWITVEG SMSEFRNPVT TLTGQTVDLE NYNEGNTYFW H AFWFAIGV ...String: HGEKSQAAFM RMRTIHWYDL SWSKEKVKIN ETVEIKGKFH VFEGWPETVD EPDVAFLNVG MPGPVFIRKE SYIGGQLVPR SVRLEIGKT YDFRVVLKAR RPGDWHVHTM MNVQGGGPII GPGKWITVEG SMSEFRNPVT TLTGQTVDLE NYNEGNTYFW H AFWFAIGV AWIGYWSRRP IFIPRLLMVD AGRADELVSA TDRKVAMGFL AATILIVVMA MSSANSKYPI TIPLQAGTMR GM KPLELPA PTVSVKVEDA TYRVPGRAMR MKLTITNHGN SPIRLGEFYT ASVRFLDSDV YKDTTGYPED LLAEDGLSVS DNS PLAPGE TRTVDVTASD AAWEVYRLSD IIYDPDSRFA GLLFFFDATG NRQVVQIDAP LIPSFM UniProtKB: Particulate methane monooxygenase alpha subunit |
-Macromolecule #2: Particulate methane monooxygenase beta subunit
| Macromolecule | Name: Particulate methane monooxygenase beta subunit / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: methane monooxygenase (particulate) |
|---|---|
| Source (natural) | Organism:  Methylococcus capsulatus (bacteria) / Strain: Bath Methylococcus capsulatus (bacteria) / Strain: Bath |
| Molecular weight | Theoretical: 28.445098 KDa |
| Sequence | String: MSAAQSAVRS HAEAVQVSRT IDWMALFVVF FVIVGSYHIH AMLTMGDWDF WSDWKDRRLW VTVTPIVLVT FPAAVQSYLW ERYRLPWGA TVCVLGLLLG EWINRYFNFW GWTYFPINFV FPASLVPGAI ILDTVLMLSG SYLFTAIVGA MGWGLIFYPG N WPIIAPLH ...String: MSAAQSAVRS HAEAVQVSRT IDWMALFVVF FVIVGSYHIH AMLTMGDWDF WSDWKDRRLW VTVTPIVLVT FPAAVQSYLW ERYRLPWGA TVCVLGLLLG EWINRYFNFW GWTYFPINFV FPASLVPGAI ILDTVLMLSG SYLFTAIVGA MGWGLIFYPG N WPIIAPLH VPVEYNGMLM SIADIQGYNY VRTGTPEYIR MVEKGTLRTF GKDVAPVSAF FSAFMSILIY FMWHFIGRWF SN ERFLQST UniProtKB: Particulate methane monooxygenase beta subunit |
-Macromolecule #3: Ammonia monooxygenase/methane monooxygenase, subunit C family protein
| Macromolecule | Name: Ammonia monooxygenase/methane monooxygenase, subunit C family protein type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methylococcus capsulatus (bacteria) / Strain: Bath Methylococcus capsulatus (bacteria) / Strain: Bath |
| Molecular weight | Theoretical: 27.65684 KDa |
| Sequence | String: LLDKKWLTFA LAIYTVFYLW VRWYEGVYGW SAGLDSFAPE FETYWMNFLY TEIVLEIVTA SILWGYLWKT RDRNLAALTP REELRRNFT HLVWLVAYAW AIYWGASYFT EQDGTWHQTI VRDTDFTPSH IIEFYLSYPI YIITGFAAFI YAKTRLPFFA K GISLPYLV ...String: LLDKKWLTFA LAIYTVFYLW VRWYEGVYGW SAGLDSFAPE FETYWMNFLY TEIVLEIVTA SILWGYLWKT RDRNLAALTP REELRRNFT HLVWLVAYAW AIYWGASYFT EQDGTWHQTI VRDTDFTPSH IIEFYLSYPI YIITGFAAFI YAKTRLPFFA K GISLPYLV LVVGPFMILP NVGLNEWGHT FWFMEELFVA PLHYGFVIFG WLALAVMGTL TQTFYSFAQG GLGQSLCE UniProtKB: Ammonia monooxygenase/methane monooxygenase, subunit C family protein |
-Macromolecule #4: COPPER (II) ION
| Macromolecule | Name: COPPER (II) ION / type: ligand / ID: 4 / Number of copies: 9 / Formula: CU |
|---|---|
| Molecular weight | Theoretical: 63.546 Da |
| Chemical component information |  ChemComp-CU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.3 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.12 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 864874 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)