+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | WT CRISPR-Cas12a with a 5bp R-loop | |||||||||
 Map data Map data | 5bp R-loop unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / R-loop / endonuclease / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationBacillus subtilis ribonuclease / deoxyribonuclease I / deoxyribonuclease I activity / defense response to virus / lyase activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |  Acidaminococcus sp. BV3L6 (bacteria) / synthetic construct (others) Acidaminococcus sp. BV3L6 (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Strohkendl I / Taylor DW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Cas12a domain flexibility guides R-loop formation and forces RuvC resetting. Authors: Isabel Strohkendl / Aakash Saha / Catherine Moy / Alexander-Hoi Nguyen / Mohd Ahsan / Rick Russell / Giulia Palermo / David W Taylor /  Abstract: The specific nature of CRISPR-Cas12a makes it a desirable RNA-guided endonuclease for biotechnology and therapeutic applications. To understand how R-loop formation within the compact Cas12a enables ...The specific nature of CRISPR-Cas12a makes it a desirable RNA-guided endonuclease for biotechnology and therapeutic applications. To understand how R-loop formation within the compact Cas12a enables target recognition and nuclease activation, we used cryo-electron microscopy to capture wild-type Acidaminococcus sp. Cas12a R-loop intermediates and DNA delivery into the RuvC active site. Stages of Cas12a R-loop formation-starting from a 5-bp seed-are marked by distinct REC domain arrangements. Dramatic domain flexibility limits contacts until nearly complete R-loop formation, when the non-target strand is pulled across the RuvC nuclease and coordinated domain docking promotes efficient cleavage. Next, substantial domain movements enable target strand repositioning into the RuvC active site. Between cleavage events, the RuvC lid conformationally resets to occlude the active site, requiring re-activation. These snapshots build a structural model depicting Cas12a DNA targeting that rationalizes observed specificity and highlights mechanistic comparisons to other class 2 effectors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40441.map.gz emd_40441.map.gz | 62.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40441-v30.xml emd-40441-v30.xml emd-40441.xml emd-40441.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40441.png emd_40441.png | 58.9 KB | ||
| Filedesc metadata |  emd-40441.cif.gz emd-40441.cif.gz | 7 KB | ||
| Others |  emd_40441_half_map_1.map.gz emd_40441_half_map_1.map.gz emd_40441_half_map_2.map.gz emd_40441_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40441 http://ftp.pdbj.org/pub/emdb/structures/EMD-40441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40441 | HTTPS FTP |
-Validation report
| Summary document |  emd_40441_validation.pdf.gz emd_40441_validation.pdf.gz | 888.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40441_full_validation.pdf.gz emd_40441_full_validation.pdf.gz | 887.8 KB | Display | |
| Data in XML |  emd_40441_validation.xml.gz emd_40441_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_40441_validation.cif.gz emd_40441_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40441 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40441 | HTTPS FTP |
-Related structure data
| Related structure data |  8sfhMC  8sfiC  8sfjC  8sflC  8sfnC  8sfoC  8sfpC  8sfqC  8sfrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 5bp R-loop unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||
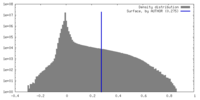
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: 5bp R-loop half map A
| File | emd_40441_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 5bp R-loop half map A | ||||||||||||
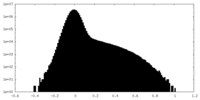
| Projections & Slices |
| ||||||||||||
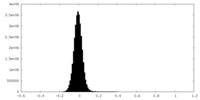
| Density Histograms |
-Half map: 5bp R-loop half map B
| File | emd_40441_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 5bp R-loop half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
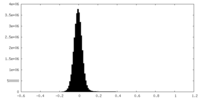
| Density Histograms |
- Sample components
Sample components
-Entire : WT AsCas12a incubated with 8bp-complementary target DNA.
| Entire | Name: WT AsCas12a incubated with 8bp-complementary target DNA. |
|---|---|
| Components |
|
-Supramolecule #1: WT AsCas12a incubated with 8bp-complementary target DNA.
| Supramolecule | Name: WT AsCas12a incubated with 8bp-complementary target DNA. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acidaminococcus sp. BV3L6 (bacteria) Acidaminococcus sp. BV3L6 (bacteria) |
-Macromolecule #1: CRISPR-associated endonuclease Cas12a
| Macromolecule | Name: CRISPR-associated endonuclease Cas12a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: deoxyribonuclease I |
|---|---|
| Source (natural) | Organism:  Acidaminococcus sp. BV3L6 (bacteria) Acidaminococcus sp. BV3L6 (bacteria) |
| Molecular weight | Theoretical: 151.418953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQFEGFTNL YQVSKTLRFE LIPQGKTLKH IQEQGFIEED KARNDHYKEL KPIIDRIYKT YADQCLQLVQ LDWENLSAAI DSYRKEKTE ETRNALIEEQ ATYRNAIHDY FIGRTDNLTD AINKRHAEIY KGLFKAELFN GKVLKQLGTV TTTEHENALL R SFDKFTTY ...String: MTQFEGFTNL YQVSKTLRFE LIPQGKTLKH IQEQGFIEED KARNDHYKEL KPIIDRIYKT YADQCLQLVQ LDWENLSAAI DSYRKEKTE ETRNALIEEQ ATYRNAIHDY FIGRTDNLTD AINKRHAEIY KGLFKAELFN GKVLKQLGTV TTTEHENALL R SFDKFTTY FSGFYENRKN VFSAEDISTA IPHRIVQDNF PKFKENCHIF TRLITAVPSL REHFENVKKA IGIFVSTSIE EV FSFPFYN QLLTQTQIDL YNQLLGGISR EAGTEKIKGL NEVLNLAIQK NDETAHIIAS LPHRFIPLFK QILSDRNTLS FIL EEFKSD EEVIQSFCKY KTLLRNENVL ETAEALFNEL NSIDLTHIFI SHKKLETISS ALCDHWDTLR NALYERRISE LTGK ITKSA KEKVQRSLKH EDINLQEIIS AAGKELSEAF KQKTSEILSH AHAALDQPLP TTLKKQEEKE ILKSQLDSLL GLYHL LDWF AVDESNEVDP EFSARLTGIK LEMEPSLSFY NKARNYATKK PYSVEKFKLN FQMPTLASGW DVNKEKNNGA ILFVKN GLY YLGIMPKQKG RYKALSFEPT EKTSEGFDKM YYDYFPDAAK MIPKCSTQLK AVTAHFQTHT TPILLSNNFI EPLEITK EI YDLNNPEKEP KKFQTAYAKK TGDQKGYREA LCKWIDFTRD FLSKYTKTTS IDLSSLRPSS QYKDLGEYYA ELNPLLYH I SFQRIAEKEI MDAVETGKLY LFQIYNKDFA KGHHGKPNLH TLYWTGLFSP ENLAKTSIKL NGQAELFYRP KSRMKRMAH RLGEKMLNKK LKDQKTPIPD TLYQELYDYV NHRLSHDLSD EARALLPNVI TKEVSHEIIK DRRFTSDKFF FHVPITLNYQ AANSPSKFN QRVNAYLKEH PETPIIGIDR GERNLIYITV IDSTGKILEQ RSLNTIQQFD YQKKLDNREK ERVAARQAWS V VGTIKDLK QGYLSQVIHE IVDLMIHYQA VVVLENLNFG FKSKRTGIAE KAVYQQFEKM LIDKLNCLVL KDYPAEKVGG VL NPYQLTD QFTSFAKMGT QSGFLFYVPA PYTSKIDPLT GFVDPFVWKT IKNHESRKHF LEGFDFLHYD VKTGDFILHF KMN RNLSFQ RGLPGFMPAW DIVFEKNETQ FDAKGTPFIA GKRIVPVIEN HRFTGRYRDL YPANELIALL EEKGIVFRDG SNIL PKLLE NDDSHAIDTM VALIRSVLQM RNSNAATGED YINSPVRDLN GVCFDSRFQN PEWPMDADAN GAYHIALKGQ LLLNH LKES KDLKLQNGIS NQDWLAYIQE LRN UniProtKB: CRISPR-associated endonuclease Cas12a |
-Macromolecule #2: RNA (27-MER)
| Macromolecule | Name: RNA (27-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.598088 KDa |
| Sequence | String: AAUUUCUACU CUUGUAGAUG UGAUAAG |
-Macromolecule #3: DNA (32-MER)
| Macromolecule | Name: DNA (32-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.882405 KDa |
| Sequence | String: (DA)(DC)(DC)(DG)(DT)(DA)(DA)(DG)(DG)(DT) (DC)(DT)(DT)(DA)(DT)(DC)(DA)(DC)(DT)(DA) (DA)(DA)(DA)(DG)(DA)(DT)(DC)(DG)(DG) (DA)(DA)(DG) |
-Macromolecule #4: DNA (32-MER)
| Macromolecule | Name: DNA (32-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.797312 KDa |
| Sequence | String: (DC)(DT)(DT)(DC)(DC)(DG)(DA)(DT)(DC)(DT) (DT)(DT)(DT)(DA)(DG)(DT)(DG)(DA)(DT)(DA) (DA)(DG)(DA)(DC)(DC)(DT)(DT)(DA)(DC) (DG)(DG)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 60595 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)