[English] 日本語
 Yorodumi
Yorodumi- EMDB-40033: Bombyx mori R2 retrotransposon initiating target-primed reverse t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bombyx mori R2 retrotransposon initiating target-primed reverse transcription | |||||||||
 Map data Map data | Sharpened map of Bombyx mori R2 retrotransposon initiating target-primed reverse transcription | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | retrotransposon / LINE / reverse transcriptase / endonuclease / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
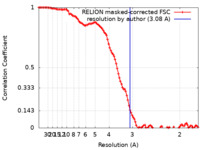
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Wilkinson ME / Zhang F | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structure of the R2 non-LTR retrotransposon initiating target-primed reverse transcription. Authors: Max E Wilkinson / Chris J Frangieh / Rhiannon K Macrae / Feng Zhang /  Abstract: Non-long terminal repeat (non-LTR) retrotransposons, or long interspersed nuclear elements (LINEs), are an abundant class of eukaryotic transposons that insert into genomes by target-primed reverse ...Non-long terminal repeat (non-LTR) retrotransposons, or long interspersed nuclear elements (LINEs), are an abundant class of eukaryotic transposons that insert into genomes by target-primed reverse transcription (TPRT). During TPRT, a target DNA sequence is nicked and primes reverse transcription of the retrotransposon RNA. Here, we report the cryo-electron microscopy structure of the R2 non-LTR retrotransposon initiating TPRT at its ribosomal DNA target. The target DNA sequence is unwound at the insertion site and recognized by an upstream motif. An extension of the reverse transcriptase (RT) domain recognizes the retrotransposon RNA and guides the 3' end into the RT active site to template reverse transcription. We used Cas9 to retarget R2 in vitro to non-native sequences, suggesting future use as a reprogrammable RNA-based gene-insertion tool. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40033.map.gz emd_40033.map.gz | 96.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40033-v30.xml emd-40033-v30.xml emd-40033.xml emd-40033.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40033_fsc.xml emd_40033_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40033.png emd_40033.png | 112.1 KB | ||
| Masks |  emd_40033_msk_1.map emd_40033_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_40033_half_map_1.map.gz emd_40033_half_map_1.map.gz emd_40033_half_map_2.map.gz emd_40033_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40033 http://ftp.pdbj.org/pub/emdb/structures/EMD-40033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40033 | HTTPS FTP |
-Related structure data
| Related structure data |  8gh6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40033.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40033.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of Bombyx mori R2 retrotransposon initiating target-primed reverse transcription | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.884 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40033_msk_1.map emd_40033_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map one
| File | emd_40033_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map one | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map two
| File | emd_40033_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map two | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Bombyx mori R2 retrotransposon initiating target-primed reverse t...
| Entire | Name: Bombyx mori R2 retrotransposon initiating target-primed reverse transcription |
|---|---|
| Components |
|
-Supramolecule #1: Bombyx mori R2 retrotransposon initiating target-primed reverse t...
| Supramolecule | Name: Bombyx mori R2 retrotransposon initiating target-primed reverse transcription type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Reverse transcriptase-like protein
| Macromolecule | Name: Reverse transcriptase-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 123.358352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMASTALSLM GRCNPDGCTR GKHVTAAPMD GPRGPSSLAG TFGWGLAIPA GEPCGRVCSP ATVGFFPVAK KSNKENRPEA SGLPLESER TGDNPTVRGS AGADPVGQDA PGWTCQFCER TFSTNRGLGV HKRRAHPVET NTDAAPMMVK RRWHGEEIDL L ARTEARLL ...String: MMASTALSLM GRCNPDGCTR GKHVTAAPMD GPRGPSSLAG TFGWGLAIPA GEPCGRVCSP ATVGFFPVAK KSNKENRPEA SGLPLESER TGDNPTVRGS AGADPVGQDA PGWTCQFCER TFSTNRGLGV HKRRAHPVET NTDAAPMMVK RRWHGEEIDL L ARTEARLL AERGQCSGGD LFGALPGFGR TLEAIKGQRR REPYRALVQA HLARFGSQPG PSSGGCSAEP DFRRASGAEE AG EERCAED AAAYDPSAVG QMSPDAARVL SELLEGAGRR RACRAMRPKT AGRRNDLHDD RTASAHKTSR QKRRAEYARV QEL YKKCRS RAAAEVIDGA CGGVGHSLEE METYWRPILE RVSDAPGPTP EALHALGRAE WHGGNRDYTQ LWKPISVEEI KASR FDWRT SPGPDGIRSG QWRAVPVHLK AEMFNAWMAR GEIPEILRQC RTVFVPKVER PGGPGEYRPI SIASIPLRHF HSILA RRLL ACCPPDARQR GFICADGTLE NSAVLDAVLG DSRKKLRECH VAVLDFAKAF DTVSHEALVE LLRLRGMPEQ FCGYIA HLY DTASTTLAVN NEMSSPVKVG RGVRQGDPLS PILFNVVMDL ILASLPERVG YRLEMELVSA LAYADDLVLL AGSKVGM QE SISAVDCVGR QMGLRLNCRK SAVLSMIPDG HRKKHHYLTE RTFNIGGKPL RQVSCVERWR YLGVDFEASG CVTLEHSI S SALNNISRAP LKPQQRLEIL RAHLIPRFQH GFVLGNISDD RLRMLDVQIR KAVGQWLRLP ADVPKAYYHA AVQDGGLAI PSVRATIPDL IVRRFGGLDS SPWSVARAAA KSDKIRKKLR WAWKQLRRFS RVDSTTQRPS VRLFWREHLH ASVDGRELRE STRTPTSTK WIRERCAQIT GRDFVQFVHT HINALPSRIR GSRGRRGGGE SSLTCRAGCK VRETTAHILQ QCHRTHGGRI L RHNKIVSF VAKAMEENKW TVELEPRLRT SVGLRKPDII ASRDGVGVIV DVQVVSGQRS LDELHREKRN KYGNHGELVE LV AGRLGLP KAECVRATSC TISWRGVWSL TSYKELRSII GLREPTLQIV PILALRGSHM NWTRFNQMTS VMGGGVG UniProtKB: Reverse transcriptase-like protein |
-Macromolecule #2: 28S DNA bottom strand, 3' side
| Macromolecule | Name: 28S DNA bottom strand, 3' side / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.507758 KDa |
| Sequence | String: (DT)(DT)(DA)(DG)(DA)(DT)(DG)(DA)(DC)(DG) (DA)(DG)(DG)(DC)(DA)(DT)(DT)(DT)(DG)(DG) (DC)(DT)(DA)(DC)(DC)(DT)(DT)(DA)(DA) (DG)(DA)(DG)(DA)(DG)(DT)(DC)(DA)(DT)(DA) (DG) (DT)(DT)(DA)(DC)(DT)(DC) ...String: (DT)(DT)(DA)(DG)(DA)(DT)(DG)(DA)(DC)(DG) (DA)(DG)(DG)(DC)(DA)(DT)(DT)(DT)(DG)(DG) (DC)(DT)(DA)(DC)(DC)(DT)(DT)(DA)(DA) (DG)(DA)(DG)(DA)(DG)(DT)(DC)(DA)(DT)(DA) (DG) (DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC) (DG)(DC)(DC)(DG)(DT)(DT)(DT)(DA)(DC)(DC) (DC)(DG) (DC)(DG)(DC)(DT)(DT)(DC)(DA) (DC)(DA)(DG) |
-Macromolecule #3: 28S DNA bottom strand, 5' side (priming strand)
| Macromolecule | Name: 28S DNA bottom strand, 5' side (priming strand) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.439812 KDa |
| Sequence | String: (DT)(DT)(DA)(DG)(DA)(DT)(DG)(DA)(DC)(DG) (DA)(DG)(DG)(DC)(DA)(DT)(DT)(DT)(DG)(DG) (DC)(DT)(DA)(DT) |
-Macromolecule #5: 28S DNA top strand
| Macromolecule | Name: 28S DNA top strand / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.654875 KDa |
| Sequence | String: (DC)(DT)(DG)(DT)(DG)(DA)(DA)(DG)(DC)(DG) (DC)(DG)(DG)(DG)(DT)(DA)(DA)(DA)(DC)(DG) (DG)(DC)(DG)(DG)(DG)(DA)(DG)(DT)(DA) (DA)(DC)(DT)(DA)(DT)(DG)(DA)(DC)(DT)(DC) (DT) (DC)(DT)(DT)(DA)(DA)(DG) ...String: (DC)(DT)(DG)(DT)(DG)(DA)(DA)(DG)(DC)(DG) (DC)(DG)(DG)(DG)(DT)(DA)(DA)(DA)(DC)(DG) (DG)(DC)(DG)(DG)(DG)(DA)(DG)(DT)(DA) (DA)(DC)(DT)(DA)(DT)(DG)(DA)(DC)(DT)(DC) (DT) (DC)(DT)(DT)(DA)(DA)(DG)(DG)(DT) (DA)(DG)(DC)(DC)(DA)(DA)(DA)(DT)(DG)(DC) (DC)(DT) (DC)(DG)(DT)(DC)(DA)(DT)(DC) (DT)(DA)(DA) |
-Macromolecule #4: R2Bm 3'UTR RNA
| Macromolecule | Name: R2Bm 3'UTR RNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.21393 KDa |
| Sequence | String: GCCUUGCACA GUAGUCCAGC GGUAAGGGUG UAGAUCAGGC CCGUCUGUUU CUCCCCCGGA GCUCGCUCCC UUGGCUUCCC UUAUAUAUU UUAACAUCAG AAACAGACAU UAAACAUCUA CUGAUCCAAU UUCGCCGGCG UACGGCCACG AUCGGGAGGG U GGGAAUCU ...String: GCCUUGCACA GUAGUCCAGC GGUAAGGGUG UAGAUCAGGC CCGUCUGUUU CUCCCCCGGA GCUCGCUCCC UUGGCUUCCC UUAUAUAUU UUAACAUCAG AAACAGACAU UAAACAUCUA CUGAUCCAAU UUCGCCGGCG UACGGCCACG AUCGGGAGGG U GGGAAUCU CGGGGGUCUU CCGAUCCUAA UCCAUGAUGA UUACGACCUG AGUCACUAAA GACGAUGGCA UGAUGAUCCG GC GAUGAAA AUAGCC |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: TTP |
|---|---|
| Molecular weight | Theoretical: 482.168 Da |
| Chemical component information |  ChemComp-TTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 Details: 20 mM HEPES-KOH pH 7.9, 500 mM potassium acetate, 5 mM magnesium acetate, 1 mM TCEP |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
| Details | A260 = 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 16551 / Average exposure time: 0.69 sec. / Average electron dose: 42.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8gh6: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)