+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
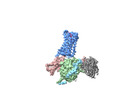
| Title | cryo-EM structure of the octreotide-bound SSTR5-Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SSTR5 / octreoitde / structural protein / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsomatostatin receptor activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / positive regulation of mini excitatory postsynaptic potential ...somatostatin receptor activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / positive regulation of mini excitatory postsynaptic potential / beta2-adrenergic receptor activity / negative regulation of smooth muscle contraction / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / AMPA selective glutamate receptor signaling pathway / Vasopressin regulates renal water homeostasis via Aquaporins / neuropeptide binding / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / positive regulation of autophagosome maturation / heat generation / norepinephrine binding / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Adrenoceptors / G alpha (q) signalling events / G alpha (i) signalling events / positive regulation of lipophagy / Thrombin signalling through proteinase activated receptors (PARs) / negative regulation of G protein-coupled receptor signaling pathway / negative regulation of multicellular organism growth / cellular response to glucocorticoid stimulus / photoreceptor outer segment membrane / response to psychosocial stress / adrenergic receptor signaling pathway / endosome to lysosome transport / spectrin binding / diet induced thermogenesis / positive regulation of cytokinesis / alkylglycerophosphoethanolamine phosphodiesterase activity / positive regulation of cAMP/PKA signal transduction / adenylate cyclase binding / smooth muscle contraction / photoreceptor outer segment / bone resorption / potassium channel regulator activity / positive regulation of bone mineralization / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / neuropeptide signaling pathway / brown fat cell differentiation / neuronal dense core vesicle / adenylate cyclase inhibitor activity / intercellular bridge / regulation of sodium ion transport / adenylate cyclase-activating adrenergic receptor signaling pathway / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / cardiac muscle cell apoptotic process / photoreceptor inner segment / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of insulin secretion / receptor-mediated endocytosis / cellular response to forskolin / response to cold / Peptide ligand-binding receptors / regulation of mitotic spindle organization / Regulation of insulin secretion / clathrin-coated endocytic vesicle membrane / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / cellular response to amyloid-beta / ADP signalling through P2Y purinoceptor 12 / mitotic spindle / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / Cargo recognition for clathrin-mediated endocytosis / GPER1 signaling / G-protein beta-subunit binding / heterotrimeric G-protein complex / glucose homeostasis / sensory perception of taste / amyloid-beta binding / positive regulation of cold-induced thermogenesis / Clathrin-mediated endocytosis Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Oplophorus gracilirostris (crustacean) / Oplophorus gracilirostris (crustacean) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Li YG / Meng XY / Yang XR / Ling SL / Shi P / Tian CL / Yang F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Acta Pharmacol Sin / Year: 2024 Journal: Acta Pharmacol Sin / Year: 2024Title: Structural insights into somatostatin receptor 5 bound with cyclic peptides. Authors: Ying-Ge Li / Xian-Yu Meng / Xiru Yang / Sheng-Long Ling / Pan Shi / Chang-Lin Tian / Fan Yang /  Abstract: Somatostatin receptor 5 (SSTR5) is highly expressed in ACTH-secreting pituitary adenomas and is an important drug target for the treatment of Cushing's disease. Two cyclic SST analog peptides ...Somatostatin receptor 5 (SSTR5) is highly expressed in ACTH-secreting pituitary adenomas and is an important drug target for the treatment of Cushing's disease. Two cyclic SST analog peptides (pasireotide and octreotide) both can activate SSTR5 and SSTR2. Pasireotide is preferential binding to SSTR5 than octreotide, while octreotide is biased to SSTR2 than SSTR5. The lack of selectivity of both pasireotide and octreotide causes side effects, such as hyperglycemia, gastrointestinal disturbance, and abnormal glucose homeostasis. However, little is known about the binding and selectivity mechanisms of pasireotide and octreotide with SSTR5, limiting the development of subtype-selective SST analog drugs specifically targeting SSTR5. Here, we report two cryo-electron microscopy (cryo-EM) structures of SSTR5-Gi complexes activated by pasireotide and octreoitde at resolutions of 3.09 Å and 3.24 Å, respectively. In combination with structural analysis and functional experiments, our results reveal the molecular mechanisms of ligand recognition and receptor activation. We also demonstrate that pasireotide preferentially binds to SSTR5 through the interactions between Tyr(Bzl)/Trp of pasireotide and SSTR5. Moreover, we find that the Q, N, F and ECL2 of SSTR2 play a crucial role in octreotide biased binding of SSTR2. Our results will provide structural insights and offer new opportunities for the drug discovery of better selective pharmaceuticals targeting specific SSTR subtypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39901.map.gz emd_39901.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39901-v30.xml emd-39901-v30.xml emd-39901.xml emd-39901.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39901.png emd_39901.png | 31.8 KB | ||
| Filedesc metadata |  emd-39901.cif.gz emd-39901.cif.gz | 6.7 KB | ||
| Others |  emd_39901_half_map_1.map.gz emd_39901_half_map_1.map.gz emd_39901_half_map_2.map.gz emd_39901_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39901 http://ftp.pdbj.org/pub/emdb/structures/EMD-39901 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39901 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39901 | HTTPS FTP |
-Related structure data
| Related structure data |  8zbeMC  8zcjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39901.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39901.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39901_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
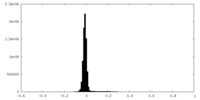
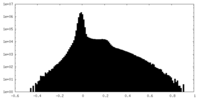
| Density Histograms |
-Half map: #1
| File | emd_39901_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
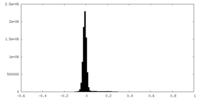
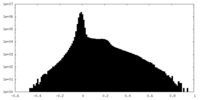
| Density Histograms |
- Sample components
Sample components
-Entire : octreotide-bound SSTR5-Gi complex
| Entire | Name: octreotide-bound SSTR5-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: octreotide-bound SSTR5-Gi complex
| Supramolecule | Name: octreotide-bound SSTR5-Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fu...
| Macromolecule | Name: Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fusion protein) type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) |
| Molecular weight | Theoretical: 62.01593 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKMGQPGN GSAFLLAPNG SHAPDHDVME PLFPASTPSW NASSPGAASG GGDNRTLVGP APSAGARAV LVPVLYLLVC AAGLGGNTLV IYVVLRFAKM KTVTNIYILN LAVADVLYML GLPFLATQNA ASFWPFGPVL C RLVMTLDG ...String: MKTIIALSYI FCLVFADYKD DDDKMGQPGN GSAFLLAPNG SHAPDHDVME PLFPASTPSW NASSPGAASG GGDNRTLVGP APSAGARAV LVPVLYLLVC AAGLGGNTLV IYVVLRFAKM KTVTNIYILN LAVADVLYML GLPFLATQNA ASFWPFGPVL C RLVMTLDG VNQFTSVFCL TVMSVDRYLA VVHPLSSARW RRPRVAKLAS AAAWVLSLCM SLPLLVFADV QEGGTCNASW PE PVGLWGA VFIIYTAVLG FFAPLLVICL CYLLIVVKVR AAGVRVGCVR RRSERKVTRM VLVVVLVFAG CWLPFFTVNI VNL AVALPQ EPASAGLYFF VVILSYANSC ANPVLYGFLS DNFRQSFQKV LCLRKGSGAK DADATEPRPD RIRQQQEATP PAHR AAANG LMQTSKLVFT LEDFVGDWEQ TAAYNLDQVL EQGGVSSLLQ NLAVSVTPIQ RIVRSGENAL KIDIHVIIPY EGLSA DQMA QIEEVFKVVY PVDDHHFKVI LPYGTLVIDG VTPNMLNYFG RPYEGIAVFD GKKITVTGTL WNGNKIIDER LITPDG SML FRVTINS UniProtKB: Beta-2 adrenergic receptor, Somatostatin receptor type 5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein subunit gamma
| Macromolecule | Name: Guanine nucleotide-binding protein subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein subunit gamma |
-Macromolecule #4: ScFv16
| Macromolecule | Name: ScFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.708473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELVD ENLYFQGASH HHHHHHH |
-Macromolecule #5: octreotide
| Macromolecule | Name: octreotide / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.036246 KDa |
| Sequence | String: (DPN)CF(DTR)KTCT |
-Macromolecule #6: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.055867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGSSGG GGSGGGGSSG VSGWRLFKKI S UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.24 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 452725 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)