+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ETAR bound with Macitentan | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / COMPLEX / ETA / MACITENTAN / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein localization to cell leading edge / endothelin receptor activity / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation / semaphorin-plexin signaling pathway involved in axon guidance / neural crest cell fate commitment / glomerular endothelium development / sympathetic neuron axon guidance / noradrenergic neuron differentiation / vascular associated smooth muscle cell development ...regulation of protein localization to cell leading edge / endothelin receptor activity / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation / semaphorin-plexin signaling pathway involved in axon guidance / neural crest cell fate commitment / glomerular endothelium development / sympathetic neuron axon guidance / noradrenergic neuron differentiation / vascular associated smooth muscle cell development / cardiac chamber formation / atrial cardiac muscle tissue development / pharyngeal arch artery morphogenesis / regulation of D-glucose transmembrane transport / endothelin receptor signaling pathway involved in heart process / cardiac neural crest cell migration involved in outflow tract morphogenesis / heparin proteoglycan metabolic process / podocyte differentiation / endothelin receptor signaling pathway / podocyte apoptotic process / positive regulation of cation channel activity / left ventricular cardiac muscle tissue morphogenesis / developmental pigmentation / embryonic skeletal system development / sodium ion homeostasis / mesenchymal cell apoptotic process / response to acetylcholine / axonogenesis involved in innervation / glomerular filtration / enteric nervous system development / renal sodium ion absorption / renal albumin absorption / cellular response to follicle-stimulating hormone stimulus / artery smooth muscle contraction / protein transmembrane transport / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / cellular response to luteinizing hormone stimulus / respiratory gaseous exchange by respiratory system / sympathetic nervous system development / vasoconstriction / cranial skeletal system development / norepinephrine metabolic process / cellular response to toxic substance / embryonic heart tube development / axon extension / cellulase / establishment of endothelial barrier / cellulase activity / aorta development / beta-glucosidase activity / middle ear morphogenesis / branching involved in blood vessel morphogenesis / neuron remodeling / neuromuscular process / face development / thyroid gland development / smooth muscle contraction / blood vessel remodeling / canonical Wnt signaling pathway / cAMP/PKA signal transduction / cellulose catabolic process / activation of adenylate cyclase activity / regulation of heart rate / Peptide ligand-binding receptors / response to amphetamine / mitochondrion organization / electron transport chain / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to wounding / calcium ion transmembrane transport / regulation of blood pressure / intracellular calcium ion homeostasis / mitotic cell cycle / positive regulation of cytosolic calcium ion concentration / cellular response to oxidative stress / gene expression / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / in utero embryonic development / response to hypoxia / electron transfer activity / periplasmic space / positive regulation of canonical NF-kappaB signal transduction / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / heme binding / cell surface / signal transduction / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
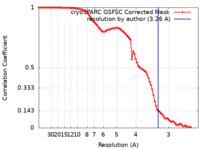
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Hou JY / Liu SH / Wu LJ / Liu ZJ / Hua T | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural basis of antagonist selectivity in endothelin receptors. Authors: Junyi Hou / Shenhui Liu / Xiaodan Zhang / Guowei Tu / Lijie Wu / Yijie Zhang / Hao Yang / Xiangcheng Li / Junlin Liu / Longquan Jiang / Qiwen Tan / Fang Bai / Zhijie Liu / Changhong Miao / Tian Hua / Zhe Luo /  Abstract: Endothelins and their receptors, ET and ET, play vital roles in maintaining vascular homeostasis. Therapeutically targeting endothelin receptors, particularly through ET antagonists, has shown ...Endothelins and their receptors, ET and ET, play vital roles in maintaining vascular homeostasis. Therapeutically targeting endothelin receptors, particularly through ET antagonists, has shown efficacy in treating pulmonary arterial hypertension (PAH) and other cardiovascular- and renal-related diseases. Here we present cryo-electron microscopy structures of ET in complex with two PAH drugs, macitentan and ambrisentan, along with zibotentan, a selective ET antagonist, respectively. Notably, a specialized anti-ET antibody facilitated the structural elucidation. These structures, together with the active-state structures of ET-1-bound ET and ET, and the agonist BQ3020-bound ET, in complex with G, unveil the molecular basis of agonist/antagonist binding modes in endothelin receptors. Key residues that confer antagonist selectivity to endothelin receptors were identified along with the activation mechanism of ET. Furthermore, our results suggest that ECL2 in ET can serve as an epitope for antibody-mediated receptor antagonism. Collectively, these insights establish a robust theoretical framework for the rational design of small-molecule drugs and antibodies with selective activity against endothelin receptors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38706.map.gz emd_38706.map.gz | 57.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38706-v30.xml emd-38706-v30.xml emd-38706.xml emd-38706.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38706_fsc.xml emd_38706_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_38706.png emd_38706.png | 62.3 KB | ||
| Filedesc metadata |  emd-38706.cif.gz emd-38706.cif.gz | 7.3 KB | ||
| Others |  emd_38706_half_map_1.map.gz emd_38706_half_map_1.map.gz emd_38706_half_map_2.map.gz emd_38706_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38706 http://ftp.pdbj.org/pub/emdb/structures/EMD-38706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38706 | HTTPS FTP |
-Related structure data
| Related structure data |  8xvjMC  8xveC  8xvhC  8xviC  8xvkC  8xvlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38706.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38706.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Endothelin receptor type A with macitentan
| Entire | Name: Complex of Endothelin receptor type A with macitentan |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Endothelin receptor type A with macitentan
| Supramolecule | Name: Complex of Endothelin receptor type A with macitentan / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1, #3-#4, #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: anti-BRIL Fab Heavy chain
| Macromolecule | Name: anti-BRIL Fab Heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.577918 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MGWSCIILFL VATATGVHSE ISEVQLVESG GGLVQPGGSL RLSCAASGFN VVDFSLHWVR QAPGKGLEWV AYISSSSGST SYADSVKGR FTISADTSKN TAYLQMNSLR AEDTAVYYCA RWGYWPGEPW WKAFDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT ...String: MGWSCIILFL VATATGVHSE ISEVQLVESG GGLVQPGGSL RLSCAASGFN VVDFSLHWVR QAPGKGLEWV AYISSSSGST SYADSVKGR FTISADTSKN TAYLQMNSLR AEDTAVYYCA RWGYWPGEPW WKAFDYWGQG TLVTVSSAST KGPSVFPLAP S SKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KV DKKVEPK SCDKTHENLY FQGHHHHHH |
-Macromolecule #2: anti-Fab Nanobody
| Macromolecule | Name: anti-Fab Nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.071431 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHGENL YFQGSQVQLQ ESGGGLVQPG GSLRLSCAAS GRTISRYAMS WFRQAPGKER EFVAVARRSG DGAFYADSVQ GRFTVSRDD AKNTVYLQMN SLKPEDTAVY YCAIDSDTFY SGSYDYWGQG TQVTVSS |
-Macromolecule #3: anti-BRIL Fab Light chain
| Macromolecule | Name: anti-BRIL Fab Light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.575604 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MGWSCIILFL VATATGVHSS DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGT DFTLTISSLQ PEDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN F YPREAKVQ ...String: MGWSCIILFL VATATGVHSS DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGT DFTLTISSLQ PEDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN F YPREAKVQ WKVDNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562
| Macromolecule | Name: Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: cellulase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.090867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHGRAMAS NYNSGLKIGA WVGTQPSESA IKSFQELQGR KLDIVHQFIN WSTDFSWVR PYADAVYNNG SILMITWEPW EYNTVDIKNG KADAYITRMA QDMKAYGKEI WLRPLHAANG DWYPWAIGYS S RVNTNETY ...String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHGRAMAS NYNSGLKIGA WVGTQPSESA IKSFQELQGR KLDIVHQFIN WSTDFSWVR PYADAVYNNG SILMITWEPW EYNTVDIKNG KADAYITRMA QDMKAYGKEI WLRPLHAANG DWYPWAIGYS S RVNTNETY IAAFRHIVDI FRANGATNVK WVFNVNCDNV GNGTSYLGHY PGDNYVDYTS IDGYNWGTTQ SWGSQWQSFD QV FSRAYQA LASINKPIII AEFASAEIGG NKARWITEAY NSIRTSYNKV IAAVWFHENK ETDWRINSSP EALAAYREAI GAE NLYFQG TTHQPTNLVL PSNGSMHNYC PQQTKITSAF KYINTVISCT IFIVGMVGNA TLLRIIYQNK CMRNGPNALI ASLA LGDLI YVVIDLPINV FKLLAGRWPF DHNDFGVFLC KLFPFLQKSS VGITVLNLCA LSVDRYRAVA SWSRVQGIGI PLVTA IEIV SIWILSFILA IPEAIGFVMV PFEYRGEQHK TCMLNATSKF MEFYQDVKDW WLFGFYFCMP LVCTAIFYTL MTCEAR RQL ADLEDNWETL NDNLKVIEKA DNAAQVKDAL TKMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALK LA NEGKVKEAQA AAEQLKTTRN AYIQKYLERA RSTLKQRREV AKTVFCLVVI FALCWFPLHL SRILKKTVYN EMDKNRCE L LSFLLLMDYI GINLATMNSC INPIALYFVS KKFKNCFQSC LCCCCYQSKS LMTSVPMNGT SI UniProtKB: Endoglucanase H, Endothelin-1 receptor, Soluble cytochrome b562, Endothelin-1 receptor |
-Macromolecule #5: Macitentan
| Macromolecule | Name: Macitentan / type: ligand / ID: 5 / Number of copies: 1 / Formula: A1D5I |
|---|---|
| Molecular weight | Theoretical: 588.273 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)