+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ETAR bound with Endothelin1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / COMPLEX / ETA / ENDOTHELIN-1 / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein localization to cell leading edge / : / endothelin A receptor binding / rhythmic excitation / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity / peptide hormone secretion / endothelin B receptor binding / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation ...regulation of protein localization to cell leading edge / : / endothelin A receptor binding / rhythmic excitation / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity / peptide hormone secretion / endothelin B receptor binding / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation / semaphorin-plexin signaling pathway involved in axon guidance / positive regulation of artery morphogenesis / histamine secretion / neural crest cell fate commitment / vein smooth muscle contraction / glomerular endothelium development / response to prostaglandin F / sympathetic neuron axon guidance / positive regulation of sarcomere organization / noradrenergic neuron differentiation / vascular associated smooth muscle cell development / positive regulation of chemokine-mediated signaling pathway / cardiac chamber formation / leukocyte activation / maternal process involved in parturition / phospholipase D-activating G protein-coupled receptor signaling pathway / rough endoplasmic reticulum lumen / atrial cardiac muscle tissue development / body fluid secretion / pharyngeal arch artery morphogenesis / regulation of D-glucose transmembrane transport / endothelin receptor signaling pathway involved in heart process / positive regulation of odontogenesis / epithelial fluid transport / cardiac neural crest cell migration involved in outflow tract morphogenesis / heparin proteoglycan metabolic process / negative regulation of hormone secretion / response to ozone / Weibel-Palade body / podocyte differentiation / endothelin receptor signaling pathway / podocyte apoptotic process / positive regulation of cation channel activity / left ventricular cardiac muscle tissue morphogenesis / developmental pigmentation / embryonic skeletal system development / positive regulation of cell growth involved in cardiac muscle cell development / sodium ion homeostasis / response to leptin / mesenchymal cell apoptotic process / response to acetylcholine / axonogenesis involved in innervation / glomerular filtration / enteric nervous system development / positive regulation of smooth muscle contraction / renal sodium ion absorption / renal albumin absorption / cellular response to follicle-stimulating hormone stimulus / artery smooth muscle contraction / positive regulation of prostaglandin secretion / protein transmembrane transport / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / cellular response to luteinizing hormone stimulus / regulation of pH / cellular response to mineralocorticoid stimulus / respiratory gaseous exchange by respiratory system / sympathetic nervous system development / vasoconstriction / basal part of cell / positive regulation of renal sodium excretion / cranial skeletal system development / response to salt / norepinephrine metabolic process / positive regulation of hormone secretion / regulation of systemic arterial blood pressure by endothelin / cellular response to toxic substance / dorsal/ventral pattern formation / embryonic heart tube development / axon extension / cellular response to fatty acid / cellulase / cartilage development / establishment of endothelial barrier / prostaglandin biosynthetic process / positive regulation of neutrophil chemotaxis / positive regulation of urine volume / signal transduction involved in regulation of gene expression / superoxide anion generation / negative regulation of protein metabolic process / cellulase activity / cellular response to glucocorticoid stimulus / nitric oxide transport / aorta development / beta-glucosidase activity / middle ear morphogenesis / branching involved in blood vessel morphogenesis / response to dexamethasone / neuron remodeling / neuromuscular process / positive regulation of cardiac muscle hypertrophy Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
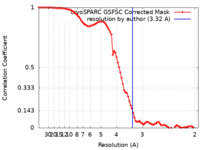
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Hou JY / Liu SH / Wu LJ / Liu ZJ / Hua T | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural basis of antagonist selectivity in endothelin receptors. Authors: Junyi Hou / Shenhui Liu / Xiaodan Zhang / Guowei Tu / Lijie Wu / Yijie Zhang / Hao Yang / Xiangcheng Li / Junlin Liu / Longquan Jiang / Qiwen Tan / Fang Bai / Zhijie Liu / Changhong Miao / Tian Hua / Zhe Luo /  Abstract: Endothelins and their receptors, ET and ET, play vital roles in maintaining vascular homeostasis. Therapeutically targeting endothelin receptors, particularly through ET antagonists, has shown ...Endothelins and their receptors, ET and ET, play vital roles in maintaining vascular homeostasis. Therapeutically targeting endothelin receptors, particularly through ET antagonists, has shown efficacy in treating pulmonary arterial hypertension (PAH) and other cardiovascular- and renal-related diseases. Here we present cryo-electron microscopy structures of ET in complex with two PAH drugs, macitentan and ambrisentan, along with zibotentan, a selective ET antagonist, respectively. Notably, a specialized anti-ET antibody facilitated the structural elucidation. These structures, together with the active-state structures of ET-1-bound ET and ET, and the agonist BQ3020-bound ET, in complex with G, unveil the molecular basis of agonist/antagonist binding modes in endothelin receptors. Key residues that confer antagonist selectivity to endothelin receptors were identified along with the activation mechanism of ET. Furthermore, our results suggest that ECL2 in ET can serve as an epitope for antibody-mediated receptor antagonism. Collectively, these insights establish a robust theoretical framework for the rational design of small-molecule drugs and antibodies with selective activity against endothelin receptors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38705.map.gz emd_38705.map.gz | 56.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38705-v30.xml emd-38705-v30.xml emd-38705.xml emd-38705.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38705_fsc.xml emd_38705_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_38705.png emd_38705.png | 53.7 KB | ||
| Filedesc metadata |  emd-38705.cif.gz emd-38705.cif.gz | 7.1 KB | ||
| Others |  emd_38705_half_map_1.map.gz emd_38705_half_map_1.map.gz emd_38705_half_map_2.map.gz emd_38705_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38705 http://ftp.pdbj.org/pub/emdb/structures/EMD-38705 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38705 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38705 | HTTPS FTP |
-Related structure data
| Related structure data |  8xviMC  8xveC  8xvhC  8xvjC  8xvkC  8xvlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38705.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38705.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Complex of protein Gs/q with ETA and Endothelin-1
| Entire | Name: Complex of protein Gs/q with ETA and Endothelin-1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of protein Gs/q with ETA and Endothelin-1
| Supramolecule | Name: Complex of protein Gs/q with ETA and Endothelin-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit...
| Macromolecule | Name: Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.464314 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHENLY FQGNSKTEDQ RNEEKAQREA NKKIEKQLQK DKQVYRATHR LLLLGADNSG KSTIVKQMRI LHGGSGGSGG TSGIFETKF QVDKVNFHMF DVGGQRDERR KWIQCFNDVT AIIFVVDSSD YNRLQEALNL FKSIWNNRWL RTISVILFLN K QDLLAEKV ...String: HHHHHHENLY FQGNSKTEDQ RNEEKAQREA NKKIEKQLQK DKQVYRATHR LLLLGADNSG KSTIVKQMRI LHGGSGGSGG TSGIFETKF QVDKVNFHMF DVGGQRDERR KWIQCFNDVT AIIFVVDSSD YNRLQEALNL FKSIWNNRWL RTISVILFLN K QDLLAEKV LAGKSKIEDY FPEFARYTTP EDATPEPGED PRVTRAKYFI RDEFLRISTA SGDGRHYCYP HFTCAVDTEN AR RIFNDCK DIILQMNLRE YNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.045629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IGRARGFSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG ...String: IGRARGFSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TGHESDINAI CF FPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADRAGVLAGH DNR VSCLGV TDDGMAVATG SWDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.057271 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMQVQLQES GGGLVQPGGS LRLSCAASGF TFSNYKMNWV RQAPGKGLEW VSDISQSGAS ISYTGSVKG RFTISRDNAK NTLYLQMNSL KPEDTAVYYC ARCPAPFTRD CFDVTSTTYA YRGQGTQVTV SSHHHHHH |
-Macromolecule #5: Endoglucanase H,Endothelin-1 receptor
| Macromolecule | Name: Endoglucanase H,Endothelin-1 receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: cellulase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.879422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAGRAMAS NYNSGLKIGA WVGTQPSESA IKSFQELQGR KLDIVHQFIN WSTDFSWVRP YADAVYNNG SILMITWEPW EYNTVDIKNG KADAYITRMA QDMKAYGKEI WLRPLHAANG DWYPWAIGYS SRVNTNETYI A AFRHIVDI ...String: MKTIIALSYI FCLVFADYKD DDDAGRAMAS NYNSGLKIGA WVGTQPSESA IKSFQELQGR KLDIVHQFIN WSTDFSWVRP YADAVYNNG SILMITWEPW EYNTVDIKNG KADAYITRMA QDMKAYGKEI WLRPLHAANG DWYPWAIGYS SRVNTNETYI A AFRHIVDI FRANGATNVK WVFNVNCDNV GNGTSYLGHY PGDNYVDYTS IDGYNWGTTQ SWGSQWQSFD QVFSRAYQAL AS INKPIII AEFASAEIGG NKARWITEAY NSIRTSYNKV IAAVWFHENK ETDWRINSSP EALAAYREAI GATTHQPTNL VLP SNGSMH NYCPQQTKIT SAFKYINTVI SCTIFIVGMV GNATLLRIIY QNKCMRNGPN ALIASLALGD LIYVVIDLPI NVFK LLAGR WPFDHNDFGV FLCKLFPFLQ KSSVGITVLN LCALSVDRYR AVASWSRVQG IGIPLVTAIE IVSIWILSFI LAIPE AIGF VMVPFEYRGE QHKTCMLNAT SKFMEFYQDV KDWWLFGFYF CMPLVCTAIF YTLMTCEMLN RRNGSLRIAL SEHLKQ RRE VAKTVFCLVV IFALCWFPLH LSRILKKTVY NEMDKNRCEL LSFLLLMDYI GINLATMNSC INPIALYFVS KKFKNCF QS CLCCCCYQSK SLMTSVPMNG TSILEVLFQG PHHHHHHHHH H UniProtKB: Endoglucanase H, Endothelin-1 receptor |
-Macromolecule #6: Endothelin-1
| Macromolecule | Name: Endothelin-1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.497951 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CSCSSLMDKE CVYFCHLDII W UniProtKB: Endothelin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)