[English] 日本語
 Yorodumi
Yorodumi- EMDB-38609: The Cryo-EM structure of IL-12, receptor subunit beta-1 and recep... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex | |||||||||
 Map data Map data | It is a density map generated in C1 symmetry. The central region has a resolution close to 3 angstrom, while the peripheral region exhibited weaker density with low resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IL-12 / IL-12RB1 / IL-12RB2 / receptor complex / CYTOKINE | |||||||||
| Function / homology |  Function and homology information Function and homology informationinterleukin-27 binding / interleukin-12 beta subunit binding / late endosome lumen / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / positive regulation of dendritic cell chemotaxis / positive regulation of natural killer cell activation / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target ...interleukin-27 binding / interleukin-12 beta subunit binding / late endosome lumen / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / positive regulation of dendritic cell chemotaxis / positive regulation of natural killer cell activation / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of vascular endothelial growth factor signaling pathway / negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of lymphocyte proliferation / positive regulation of tissue remodeling / positive regulation of T-helper 1 type immune response / sexual reproduction / positive regulation of NK T cell activation / positive regulation of smooth muscle cell apoptotic process / positive regulation of natural killer cell mediated cytotoxicity / positive regulation of mononuclear cell proliferation / interleukin-23-mediated signaling pathway / interleukin-12 receptor binding / interleukin-12 receptor complex / positive regulation of memory T cell differentiation / T-helper cell differentiation / interleukin-23 receptor complex / natural killer cell activation / Interleukin-23 signaling / positive regulation of T-helper 17 type immune response / positive regulation of osteoclast differentiation / interleukin-12-mediated signaling pathway / negative regulation of interleukin-17 production / positive regulation of NK T cell proliferation / Interleukin-12 signaling / Interleukin-35 Signalling / response to UV-B / positive regulation of natural killer cell proliferation / positive regulation of granulocyte macrophage colony-stimulating factor production / cytokine receptor activity / T-helper 1 type immune response / negative regulation of interleukin-10 production / defense response to protozoan / positive regulation of activated T cell proliferation / cytokine binding / positive regulation of interleukin-17 production / Interleukin-10 signaling / cell surface receptor signaling pathway via JAK-STAT / positive regulation of interleukin-10 production / negative regulation of protein secretion / positive regulation of defense response to virus by host / coreceptor activity / positive regulation of T-helper 17 cell lineage commitment / positive regulation of T cell proliferation / T cell proliferation / positive regulation of tyrosine phosphorylation of STAT protein / extrinsic apoptotic signaling pathway / regulation of cytokine production / positive regulation of interleukin-12 production / positive regulation of cell adhesion / cytokine activity / negative regulation of inflammatory response to antigenic stimulus / negative regulation of smooth muscle cell proliferation / growth factor activity / response to virus / cellular response to virus / cellular response to type II interferon / cytokine-mediated signaling pathway / positive regulation of inflammatory response / positive regulation of T cell mediated cytotoxicity / positive regulation of non-canonical NF-kappaB signal transduction / positive regulation of type II interferon production / positive regulation of tumor necrosis factor production / cell migration / cellular response to lipopolysaccharide / Interleukin-4 and Interleukin-13 signaling / defense response to virus / defense response to Gram-negative bacterium / response to lipopolysaccharide / receptor complex / cell surface receptor signaling pathway / defense response to Gram-positive bacterium / immune response / protein heterodimerization activity / endoplasmic reticulum lumen / external side of plasma membrane / positive regulation of cell population proliferation / protein-containing complex binding / protein kinase binding / signal transduction / extracellular space / extracellular region / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Chen HQ / Wang XQ / Ge XF / Zeng JW / Wang JW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Structure and assembly of the human IL-12 signaling complex. Authors: Huiqin Chen / Xiaofei Ge / Chun Li / Jianwei Zeng / Xinquan Wang /  Abstract: Interleukin (IL)-12 is a heterodimeric pro-inflammatory cytokine. Our cryoelectron microscopy structure determination of human IL-12 in complex with IL-12Rβ1 and IL-12Rβ2 at a resolution of 3. ...Interleukin (IL)-12 is a heterodimeric pro-inflammatory cytokine. Our cryoelectron microscopy structure determination of human IL-12 in complex with IL-12Rβ1 and IL-12Rβ2 at a resolution of 3.75 Å reveals that IL-12Rβ2 primarily interacts with the IL-12p35 subunit via its N-terminal Ig-like domain, while IL-12Rβ1 binds to the p40 subunit with its N-terminal fibronectin III domain. This binding mode of IL-12 with its receptors is similar to that of IL-23 but shows notable differences with other cytokines. Through structural information and biochemical assays, we identified Y62, Y189, and K192 as key residues in IL-12p35, which bind to IL-12Rβ2 with high affinity and mediate IL-12 signal transduction. Furthermore, structural comparisons reveal two distinctive conformational states and structural plasticity of the heterodimeric interface in IL-12. As a result, our study advances our understanding of IL-12 signal initiation and opens up new opportunities for the engineering and therapeutic targeting of IL-12. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38609.map.gz emd_38609.map.gz | 398.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38609-v30.xml emd-38609-v30.xml emd-38609.xml emd-38609.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38609.png emd_38609.png | 110.3 KB | ||
| Filedesc metadata |  emd-38609.cif.gz emd-38609.cif.gz | 6.5 KB | ||
| Others |  emd_38609_half_map_1.map.gz emd_38609_half_map_1.map.gz emd_38609_half_map_2.map.gz emd_38609_half_map_2.map.gz | 391.1 MB 391.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38609 http://ftp.pdbj.org/pub/emdb/structures/EMD-38609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38609 | HTTPS FTP |
-Validation report
| Summary document |  emd_38609_validation.pdf.gz emd_38609_validation.pdf.gz | 999.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38609_full_validation.pdf.gz emd_38609_full_validation.pdf.gz | 999.1 KB | Display | |
| Data in XML |  emd_38609_validation.xml.gz emd_38609_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_38609_validation.cif.gz emd_38609_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38609 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38609 | HTTPS FTP |
-Related structure data
| Related structure data |  8xrpMC  8yi7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38609.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38609.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
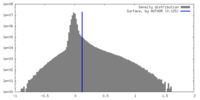
| Annotation | It is a density map generated in C1 symmetry. The central region has a resolution close to 3 angstrom, while the peripheral region exhibited weaker density with low resolution. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8697 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: map half A
| File | emd_38609_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
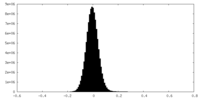
| Annotation | map_half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: map half B
| File | emd_38609_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
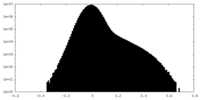
| Annotation | map_half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The Cryo-EM structure of IL-12, receptor subunit beta-1 and recep...
| Entire | Name: The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex |
|---|---|
| Components |
|
-Supramolecule #1: The Cryo-EM structure of IL-12, receptor subunit beta-1 and recep...
| Supramolecule | Name: The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 485 KDa |
-Macromolecule #1: Interleukin-12 subunit alpha
| Macromolecule | Name: Interleukin-12 subunit alpha / type: protein_or_peptide / ID: 1 / Details: Interleukin-12 subunit alpha / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.781508 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: LSLARNLPVA TPDPGMFPCL HHSQNLLRAV SNMLQKARQT LEFYPCTSEE IDHEDITKDK TSTVEACLPL ELTKNESCLN SRETSFITN GSCLASRKTS FMMALCLSSI YEDLKMYQVE FKTMNAKLLM DPKRQIFLDQ NMLAVIDELM QALNFNSETV P QKSSLEEP ...String: LSLARNLPVA TPDPGMFPCL HHSQNLLRAV SNMLQKARQT LEFYPCTSEE IDHEDITKDK TSTVEACLPL ELTKNESCLN SRETSFITN GSCLASRKTS FMMALCLSSI YEDLKMYQVE FKTMNAKLLM DPKRQIFLDQ NMLAVIDELM QALNFNSETV P QKSSLEEP DFYKTKIKLC ILLHAFRIRA VTIDRVMSYL NASHHHHHH UniProtKB: Interleukin-12 subunit alpha |
-Macromolecule #2: Interleukin-12 subunit beta
| Macromolecule | Name: Interleukin-12 subunit beta / type: protein_or_peptide / ID: 2 / Details: Interleukin-12 subunit beta / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.811023 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: AIWELKKDVY VVELDWYPDA PGEMVVLTCD TPEEDGITWT LDQSSEVLGS GKTLTIQVKE FGDAGQYTCH KGGEVLSHSL LLLHKKEDG IWSTDILKDQ KEPKNKTFLR CEAKNYSGRF TCWWLTTIST DLTFSVKSSR GSSDPQGVTC GAATLSAERV R GDNKEYEY ...String: AIWELKKDVY VVELDWYPDA PGEMVVLTCD TPEEDGITWT LDQSSEVLGS GKTLTIQVKE FGDAGQYTCH KGGEVLSHSL LLLHKKEDG IWSTDILKDQ KEPKNKTFLR CEAKNYSGRF TCWWLTTIST DLTFSVKSSR GSSDPQGVTC GAATLSAERV R GDNKEYEY SVECQEDSAC PAAEESLPIE VMVDAVHKLK YENYTSSFFI RDIIKPDPPK NLQLKPLKNS RQVEVSWEYP DT WSTPHSY FSLTFCVQVQ GKSKREKKDR VFTDKTSATV ICRKNASISV RAQDRYYSSS WSEWASVPCS UniProtKB: Interleukin-12 subunit beta |
-Macromolecule #3: Interleukin-12 receptor subunit beta-2
| Macromolecule | Name: Interleukin-12 receptor subunit beta-2 / type: protein_or_peptide / ID: 3 / Details: Interleukin-12 receptor subunit beta-2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.627477 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: KIDACKRGDV TVKPSHVILL GSTVNITCSL KPRQGCFHYS RRNKLILYKF DRRINFHHGH SLNSQVTGLP LGTTLFVCKL ACINSDEIQ ICGAEIFVGV APEQPQNLSC IQKGEQGTVA CTWERGRDTH LYTEYTLQLS GPKNLTWQKQ CKDIYCDYLD F GINLTPES ...String: KIDACKRGDV TVKPSHVILL GSTVNITCSL KPRQGCFHYS RRNKLILYKF DRRINFHHGH SLNSQVTGLP LGTTLFVCKL ACINSDEIQ ICGAEIFVGV APEQPQNLSC IQKGEQGTVA CTWERGRDTH LYTEYTLQLS GPKNLTWQKQ CKDIYCDYLD F GINLTPES PESNFTAKVT AVNSLGSSSS LPSTFTFLDI VRPLPPWDIR IKFQKASVSR CTLYWRDEGL VLLNRLRYRP SN SRLWNMV NVTKAKGRHD LLDLKPFTEY EFQISSKLHL YKGSWSDWSE SLRAQTPEEH HHHHH UniProtKB: Interleukin-12 receptor subunit beta-2 |
-Macromolecule #4: Interleukin-12 receptor subunit beta-1
| Macromolecule | Name: Interleukin-12 receptor subunit beta-1 / type: protein_or_peptide / ID: 4 / Details: Interleukin-12 receptor subunit beta-1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.736449 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD ...String: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD DTESCLCPLE MNVAQEFQLR RRQLGSQGSS WSKWSSPVCV PPENPHHHHH H UniProtKB: Interleukin-12 receptor subunit beta-1 |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.34 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 25mM Tris-HCl pH 8.0,150mM NaCl | |||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)