[English] 日本語
 Yorodumi
Yorodumi- EMDB-38565: Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP protomer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the sea urchin spSLC9C1 in state-2 w/o cAMP protomer | |||||||||
 Map data Map data | map of spSLC9C1 state-2 protomer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sodium-hydrogen exchanger / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsperm head / potassium:proton antiporter activity / sodium:proton antiporter activity / sodium ion import across plasma membrane / cGMP binding / single fertilization / sperm flagellum / cAMP binding / potassium ion transmembrane transport / regulation of intracellular pH ...sperm head / potassium:proton antiporter activity / sodium:proton antiporter activity / sodium ion import across plasma membrane / cGMP binding / single fertilization / sperm flagellum / cAMP binding / potassium ion transmembrane transport / regulation of intracellular pH / protein homodimerization activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Qu H / Zheng X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Insight / Year: 2024 Journal: Cell Insight / Year: 2024Title: Structures of a sperm-specific sodium-hydrogen exchanger. Authors: Hongyuan Qu / Yi Zhen / Mohan Xu / Yan Huang / Yashu Wang / Gaoyuan Ji / Yuyu Zhang / Haitao Li / Zigang Dong / Xiangdong Zheng /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38565.map.gz emd_38565.map.gz | 57.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38565-v30.xml emd-38565-v30.xml emd-38565.xml emd-38565.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38565.png emd_38565.png | 122.6 KB | ||
| Filedesc metadata |  emd-38565.cif.gz emd-38565.cif.gz | 6.2 KB | ||
| Others |  emd_38565_half_map_1.map.gz emd_38565_half_map_1.map.gz emd_38565_half_map_2.map.gz emd_38565_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38565 http://ftp.pdbj.org/pub/emdb/structures/EMD-38565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38565 | HTTPS FTP |
-Related structure data
| Related structure data |  8xq4MC  8xpqC  8xq7C  8xq8C  8xq9C  8xqaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38565.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38565.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of spSLC9C1 state-2 protomer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
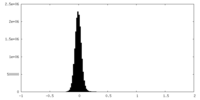
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: map of spSLC9C1 state-2 protomer
| File | emd_38565_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of spSLC9C1 state-2 protomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
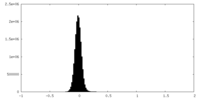
| Density Histograms |
-Half map: half map B
| File | emd_38565_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
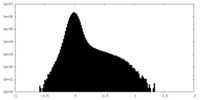
| Density Histograms |
- Sample components
Sample components
-Entire : solute carrier 9C1 protomer
| Entire | Name: solute carrier 9C1 protomer |
|---|---|
| Components |
|
-Supramolecule #1: solute carrier 9C1 protomer
| Supramolecule | Name: solute carrier 9C1 protomer / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sperm-specific sodium proton exchanger
| Macromolecule | Name: Sperm-specific sodium proton exchanger / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 144.950125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GSSSGGSGHT PTTQATHADD HDLTTHNGTE EHDDGHDDGH DDLHAHAPKV IVFISGSCLF GAISRSLFKK LPIPYTVVLL ILGAILGVV ASNVPLVEEH TRDVAHMDPH VLLQIFLPVL IFESAFAMDV HTFMRSFSQV CILALFGLVV ASVLTAVLAM N LFNYNWNF ...String: GSSSGGSGHT PTTQATHADD HDLTTHNGTE EHDDGHDDGH DDLHAHAPKV IVFISGSCLF GAISRSLFKK LPIPYTVVLL ILGAILGVV ASNVPLVEEH TRDVAHMDPH VLLQIFLPVL IFESAFAMDV HTFMRSFSQV CILALFGLVV ASVLTAVLAM N LFNYNWNF SEAMMFGAIM SATDPVAVVA LLKDLGASKQ LGTIIEGESL LNDGCAIVIF NVFMKMVFFP QLTSTVGQNV LY FLQVAVA GPLWGYAVAK VTVFFLSHIF NDALVEITIT LAATYLTYYI GDIWLEVSGV LAVVVLGLIV NAEKTSISPE VEV FLHRFW EMLAYLANTL IFMMVGVVVT QKALVAVDKM DWFYLIILYL AITIIRGMVI SLFSPILSRI GYGLTWRNAV IMTW GGLRG AVGLALALVV ENLAGNDVIG SKFLFHTAGI VVLTLVINAT TIQTLLRILG MSDISIPKRL AMAGAVRRIH EGQNR TLNM LKSDRFLADA DWDIATAACE ISDPYSALSD DENAPADELT LGERKSVCPG CKAMVPNEPS PREFADMMEE ARLRML KAE KISYWKQFEH GMLAREALRL LVQHAEVAAD EKDQFILVDD LKKSWQIKGI YPWLKRKLED LISEKKIAAI PMPKYKL GK LMYKICHHMA FEVTINIAIV LNIVPIIMEF VVQDKMASVS TMAAPGSTVS SEPSSLQKIE DALRISNYVF FVIYAIEA I VKILGLGRHY IVSHWNKFDA FILVVALVDI IIAETLLKGS ITINLSSIKV VKLFRLLRGL RMLRLTKALI PKLILVVNG KINNQLSLGY DVGKGYIIGE EEVGKIIDRM VDNKKILREL KHISETGRLQ VVKELGLLQR EHPGIAVSVK TRQAIRTILN HSRETIHEL QGAGLLDEME AHKLELTVEI KMKRLMNAPS SIPPPPPENL LKNVSWLAGD MKLIDFIKAR ASLLHFDYGE V IVREGDES DGLFLIVSGL VKLYGKSAFL DHDNPPVTAG SEENEVFEDY LTVGNVIGEM GVLTKKPRNA TVTCETTVQV YF ITAEDMN IAIDTFTLYP SLEYRLWRVV AIRIATPLIM EQMAFQGWTQ EKVKLHLERG YLVDLAESHF QFNIDATLED VIL INGTAY NAHTREEIRS PCLISRTVHK LTFQYTATEE PRLFVVRNAE YNGPILDGRL DVDSKRSLIS ITEISSNMCL KHAA ELRQK NSKVMLSRKS SGAAAKEEED CIPNTSDVEQ AAGVSPSVPT KTTPKPKSFL PSLGLSMSKE RVNGEAVEES PVKTK QGEE TPETEEGAAP RVNVASNSLE VLFQ UniProtKB: Sperm-specific sodium:proton exchanger |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)