[English] 日本語
 Yorodumi
Yorodumi- EMDB-38439: Structure of human 3-methylcrotonyl-CoA carboxylase at apo-state ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Biotin-dependent carboxylase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology information3-Methylcrotonyl-CoA carboxylase deficiency / methylcrotonoyl-CoA carboxylase / biotin metabolic process / methylcrotonoyl-CoA carboxylase complex / methylcrotonoyl-CoA carboxylase activity / L-leucine catabolic process / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / biotin carboxylase activity / branched-chain amino acid catabolic process ...3-Methylcrotonyl-CoA carboxylase deficiency / methylcrotonoyl-CoA carboxylase / biotin metabolic process / methylcrotonoyl-CoA carboxylase complex / methylcrotonoyl-CoA carboxylase activity / L-leucine catabolic process / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / biotin carboxylase activity / branched-chain amino acid catabolic process / Branched-chain amino acid catabolism / coenzyme A metabolic process / biotin binding / mitochondrial matrix / mitochondrion / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.29 Å | |||||||||
 Authors Authors | Zhou FY / Zhang YY / Zhou Q / Hu Q | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC) Authors: Zhou F / Zhang Y / Zhu Y / Zhou Q / Shi Y / Hu Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38439.map.gz emd_38439.map.gz | 419.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38439-v30.xml emd-38439-v30.xml emd-38439.xml emd-38439.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38439.png emd_38439.png | 31.9 KB | ||
| Filedesc metadata |  emd-38439.cif.gz emd-38439.cif.gz | 5.8 KB | ||
| Others |  emd_38439_half_map_1.map.gz emd_38439_half_map_1.map.gz emd_38439_half_map_2.map.gz emd_38439_half_map_2.map.gz | 412 MB 412 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38439 http://ftp.pdbj.org/pub/emdb/structures/EMD-38439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38439 | HTTPS FTP |
-Related structure data
| Related structure data |  8xl6MC  8xl3C  8xl4C  8xl5C  8xl7C  8xl8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38439.map.gz / Format: CCP4 / Size: 443.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38439.map.gz / Format: CCP4 / Size: 443.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38439_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38439_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterododecamer complex of human 3-methylcrotonyl-CoA carboxylase...
| Entire | Name: Heterododecamer complex of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) |
|---|---|
| Components |
|
-Supramolecule #1: Heterododecamer complex of human 3-methylcrotonyl-CoA carboxylase...
| Supramolecule | Name: Heterododecamer complex of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial
| Macromolecule | Name: Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: methylcrotonoyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.584016 KDa |
| Sequence | String: MAAASAVSVL LVAAERNRWH RLPSLLLPPR TWVWRQRTMK YTTATGRNIT KVLIANRGEI ACRVMRTAKK LGVQTVAVYS EADRNSMHV DMADEAYSIG PAPSQQSYLS MEKIIQVAKT SAAQAIHPGC GFLSENMEFA ELCKQEGIIF IGPPPSAIRD M GIKSTSKS ...String: MAAASAVSVL LVAAERNRWH RLPSLLLPPR TWVWRQRTMK YTTATGRNIT KVLIANRGEI ACRVMRTAKK LGVQTVAVYS EADRNSMHV DMADEAYSIG PAPSQQSYLS MEKIIQVAKT SAAQAIHPGC GFLSENMEFA ELCKQEGIIF IGPPPSAIRD M GIKSTSKS IMAAAGVPVV EGYHGEDQSD QCLKEHARRI GYPVMIKAVR GGGGKGMRIV RSEQEFQEQL ESARREAKKS FN DDAMLIE KFVDTPRHVE VQVFGDHHGN AVYLFERDCS VQRRHQKIIE EAPAPGIKSE VRKKLGEAAV RAAKAVNYVG AGT VEFIMD SKHNFCFMEM NTRLQVEHPV TEMITGTDLV EWQLRIAAGE KIPLSQEEIT LQGHAFEARI YAEDPSNNFM PVAG PLVHL STPRADPSTR IETGVRQGDE VSVHYDPMIA KLVVWAADRQ AALTKLRYSL RQYNIVGLHT NIDFLLNLSG HPEFE AGNV HTDFIPQHHK QLLLSRKAAA KESLCQAALG LILKEKAMTD TFTLQAHDQF SPFSSSSGRR LNISYTRNMT LKDGKN NVA IAVTYNHDGS YSMQIEDKTF QVLGNLYSEG DCTYLKCSVN GVASKAKLII LENTIYLFSK EGSIEIDIPV PKYLSSV SS QETQGGPLAP MTGTIEKVFV KAGDKVKAGD SLMVMIAMKM EHTIKSPKDG TVKKVFYREG AQANRHTPLV EFEEEESD K RESE UniProtKB: Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial |
-Macromolecule #2: Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial
| Macromolecule | Name: Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: methylcrotonoyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.406027 KDa |
| Sequence | String: MWAVLRLALR PCARASPAGP RAYHGDSVAS LGTQPDLGSA LYQENYKQMK ALVNQLHERV EHIKLGGGEK ARALHISRGK LLPRERIDN LIDPGSPFLE LSQFAGYQLY DNEEVPGGGI ITGIGRVSGV ECMIIANDAT VKGGAYYPVT VKKQLRAQEI A MQNRLPCI ...String: MWAVLRLALR PCARASPAGP RAYHGDSVAS LGTQPDLGSA LYQENYKQMK ALVNQLHERV EHIKLGGGEK ARALHISRGK LLPRERIDN LIDPGSPFLE LSQFAGYQLY DNEEVPGGGI ITGIGRVSGV ECMIIANDAT VKGGAYYPVT VKKQLRAQEI A MQNRLPCI YLVDSGGAYL PRQADVFPDR DHFGRTFYNQ AIMSSKNIAQ IAVVMGSCTA GGAYVPAMAD ENIIVRKQGT IF LAGPPLV KAATGEEVSA EDLGGADLHC RKSGVSDHWA LDDHHALHLT RKVVRNLNYQ KKLDVTIEPS EEPLFPADEL YGI VGANLK RSFDVREVIA RIVDGSRFTE FKAFYGDTLV TGFARIFGYP VGIVGNNGVL FSESAKKGTH FVQLCCQRNI PLLF LQNIT GFMVGREYEA EGIAKDGAKM VAAVACAQVP KITLIIGGSY GAGNYGMCGR AYSPRFLYIW PNARISVMGG EQAAN VLAT ITKDQRAREG KQFSSADEAA LKEPIIKKFE EEGNPYYSSA RVWDDGIIDP ADTRLVLGLS FSAALNAPIE KTDFGI FRM UniProtKB: Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial |
-Macromolecule #3: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 3 / Number of copies: 6 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 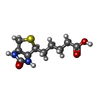 ChemComp-BTN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.29 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 407407 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































