+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a bacterial protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial immune proteins / IMMUNE SYSTEM | |||||||||
| Biological species |  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) | |||||||||
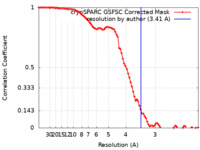
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Yu G / Liao F / Li X / Li Z / Zhang H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of a bacterial protein Authors: Yu G / Liao F / Li X / Li Z / Zhang H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37569.map.gz emd_37569.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37569-v30.xml emd-37569-v30.xml emd-37569.xml emd-37569.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37569_fsc.xml emd_37569_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_37569.png emd_37569.png | 129.7 KB | ||
| Filedesc metadata |  emd-37569.cif.gz emd-37569.cif.gz | 6.3 KB | ||
| Others |  emd_37569_half_map_1.map.gz emd_37569_half_map_1.map.gz emd_37569_half_map_2.map.gz emd_37569_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37569 http://ftp.pdbj.org/pub/emdb/structures/EMD-37569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37569 | HTTPS FTP |
-Related structure data
| Related structure data |  8wivMC  8wj3C  8wk0C  8wocC  8wodC  8wofC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37569.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37569.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07578 Å | ||||||||||||||||||||||||||||||||||||
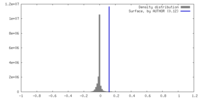
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_37569_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
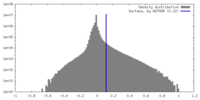
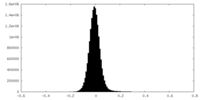
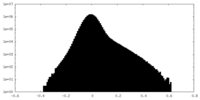
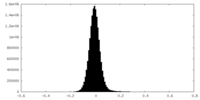
| Density Histograms |
-Half map: half map
| File | emd_37569_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
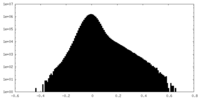
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of a bacterial protein
| Entire | Name: Cryo-EM structure of a bacterial protein |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of a bacterial protein
| Supramolecule | Name: Cryo-EM structure of a bacterial protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Helicase HerA central domain-containing protein
| Macromolecule | Name: Helicase HerA central domain-containing protein / type: protein_or_peptide / ID: 1 / Details: WP_091160506.1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) |
| Molecular weight | Theoretical: 77.626344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIGVNRMTEA STYIGTVQDV NGANIRVVLD INTISSLKFV DGQGYRIGQI GSFVRIPIGY INLFGIVSQV GAGAVPDKLL EVEPYGHRW ISVQLVGEEG IKKEFERGVS QYPTIGDKVH IVTEPDLKKI YGTQNKKYIS LGNIASVDSI PALVNIDTLV T RHSAVLGS ...String: MIGVNRMTEA STYIGTVQDV NGANIRVVLD INTISSLKFV DGQGYRIGQI GSFVRIPIGY INLFGIVSQV GAGAVPDKLL EVEPYGHRW ISVQLVGEEG IKKEFERGVS QYPTIGDKVH IVTEPDLKKI YGTQNKKYIS LGNIASVDSI PALVNIDTLV T RHSAVLGS TGSGKSTTVT SILQRISDMS QFPSARIIVF DIHGEYAAAF KGKAKVYKVT PSNNELKLSI PYWALTCDEF LS VAFGGLE GSGRNALIDK IYELKLQTLK RQEYEGINED SLTVDTPIPF SIHKLWFDLY RAEISTHYVQ GSHSEENEAL LLG EDGNPV QKGDSLKVVP PIYMPHTQAQ GATKIYLSNR GKNIRKPLEG LASLLKDPRY EFLFNADDWS VNLDGKTNKD LDAL LETWV GSEESISIFD LSGMPSSILD TLIGILIRIL YDSLFWSRNQ PEGGRERPLL VVLEEAHTYL GKDSRGIAID GVRKI VKEG RKYGIGMMLV SQRPSEIDST ILSQCGTLFA LRMNNSSDRN HVLGAVSDSF EGLMGMLPTL RTGEAIIIGE SVRLPM RTI ISPPPFGRRP DSLDPDVTAK WSNNRVQGDY KEVLTLWRQK KVRSQRIVEN IKRLPVVNEG EMTDMVREMV TSSNILS IG YEADSMTLEI EFNHGLVYQY YDVPETLHTE LLAAESHGKF FNSQIKNNYR FSRI |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 4 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)