登録情報 データベース : EMDB / ID : EMD-37365タイトル Cryo-EM structure of the Rpd3S-nucleosome complex from budding yeast in State 1 複合体 : The Rpd3S complexタンパク質・ペプチド : x 5種DNA : x 2種リガンド : x 2種 / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
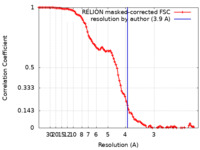
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae (パン酵母) / Homo sapiens (ヒト)手法 / / 解像度 : 3.9 Å Wang C / Zhan X 資金援助 Organization Grant number 国 National Natural Science Foundation of China (NSFC)
ジャーナル : Sci Adv / 年 : 2024タイトル : Structures and dynamics of Rpd3S complex bound to nucleosome.著者 : Chengcheng Wang / Chen Chu / Zhouyan Guo / Xiechao Zhan / 要旨 : The Rpd3S complex plays a pivotal role in facilitating local histone deacetylation in the transcribed regions to suppress intragenic transcription initiation. Here, we present the cryo-electron ... The Rpd3S complex plays a pivotal role in facilitating local histone deacetylation in the transcribed regions to suppress intragenic transcription initiation. Here, we present the cryo-electron microscopy structures of the budding yeast Rpd3S complex in both its apo and three nucleosome-bound states at atomic resolutions, revealing the exquisite architecture of Rpd3S to well accommodate a mononucleosome without linker DNA. The Rpd3S core, containing a Sin3 Lobe and two NB modules, is a rigid complex and provides three positive-charged anchors (Sin3_HCR and two Rco1_NIDs) to connect nucleosomal DNA. In three nucleosome-bound states, the Rpd3S core exhibits three distinct orientations relative to the nucleosome, assisting the sector-shaped deacetylase Rpd3 to locate above the SHL5-6, SHL0-1, or SHL2-3, respectively. Our work provides a structural framework that reveals a dynamic working model for the Rpd3S complex to engage diverse deacetylation sites. 履歴 登録 2023年9月5日 - ヘッダ(付随情報) 公開 2024年5月15日 - マップ公開 2024年5月15日 - 更新 2024年5月15日 - 現状 2024年5月15日 処理サイト : PDBj / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 中国, 1件
中国, 1件  引用
引用 ジャーナル: Sci Adv / 年: 2024
ジャーナル: Sci Adv / 年: 2024
 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_37365.map.gz
emd_37365.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-37365-v30.xml
emd-37365-v30.xml emd-37365.xml
emd-37365.xml EMDBヘッダ
EMDBヘッダ emd_37365_fsc.xml
emd_37365_fsc.xml FSCデータファイル
FSCデータファイル emd_37365.png
emd_37365.png emd-37365.cif.gz
emd-37365.cif.gz emd_37365_half_map_1.map.gz
emd_37365_half_map_1.map.gz emd_37365_half_map_2.map.gz
emd_37365_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-37365
http://ftp.pdbj.org/pub/emdb/structures/EMD-37365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37365
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37365 emd_37365_validation.pdf.gz
emd_37365_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_37365_full_validation.pdf.gz
emd_37365_full_validation.pdf.gz emd_37365_validation.xml.gz
emd_37365_validation.xml.gz emd_37365_validation.cif.gz
emd_37365_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37365
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37365 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37365
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37365 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_37365.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_37365.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)