+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of EBV gH/gL-gp42 in complex with fab 2C1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human gammaherpesvirus 4 / Neutralizing antibody / VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbohydrate binding / host cell endosome / host cell Golgi apparatus / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) / Epstein-Barr virus (strain GD1) (Epstein-Barr virus) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
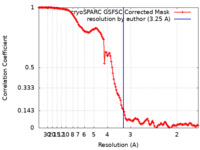
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Fang XY / Zhao GX / Zeng MS / Liu Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2024 Journal: Cell Rep Med / Year: 2024Title: Potent human monoclonal antibodies targeting Epstein-Barr virus gp42 reveal vulnerable sites for virus infection. Authors: Ge-Xin Zhao / Xin-Yan Fang / Guo-Long Bu / Shuai-Jia-Bin Chen / Cong Sun / Ting Li / Chu Xie / Yu Wang / Shu-Xin Li / Ning Meng / Guo-Kai Feng / Qian Zhong / Xiang-Wei Kong / Zheng Liu / Mu-Sheng Zeng /  Abstract: Epstein-Barr virus (EBV) is linked to various malignancies and autoimmune diseases, posing a significant global health challenge due to the lack of specific treatments or vaccines. Despite its ...Epstein-Barr virus (EBV) is linked to various malignancies and autoimmune diseases, posing a significant global health challenge due to the lack of specific treatments or vaccines. Despite its crucial role in EBV infection in B cells, the mechanisms of the glycoprotein gp42 remain elusive. In this study, we construct an antibody phage library from 100 EBV-positive individuals, leading to the identification of two human monoclonal antibodies, 2B7 and 2C1. These antibodies effectively neutralize EBV infection in vitro and in vivo while preserving gp42's interaction with the human leukocyte antigen class II (HLA-II) receptor. Structural analysis unveils their distinct binding epitopes on gp42, different from the HLA-II binding site. Furthermore, both 2B7 and 2C1 demonstrate potent neutralization of EBV infection in HLA-II-positive epithelial cells, expanding our understanding of gp42's role. Overall, this study introduces two human anti-gp42 antibodies with potential implications for developing EBV vaccines targeting gp42 epitopes, addressing a critical gap in EBV research. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37249.map.gz emd_37249.map.gz | 189.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37249-v30.xml emd-37249-v30.xml emd-37249.xml emd-37249.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37249_fsc.xml emd_37249_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_37249.png emd_37249.png | 54.4 KB | ||
| Filedesc metadata |  emd-37249.cif.gz emd-37249.cif.gz | 6.4 KB | ||
| Others |  emd_37249_half_map_1.map.gz emd_37249_half_map_1.map.gz emd_37249_half_map_2.map.gz emd_37249_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37249 http://ftp.pdbj.org/pub/emdb/structures/EMD-37249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37249 | HTTPS FTP |
-Related structure data
| Related structure data |  8khrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37249.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37249.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
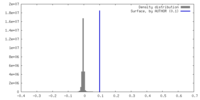
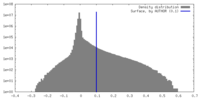
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37249_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37249_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentamer complex of gH/gL-gp42 with fab 2C1
| Entire | Name: Pentamer complex of gH/gL-gp42 with fab 2C1 |
|---|---|
| Components |
|
-Supramolecule #1: Pentamer complex of gH/gL-gp42 with fab 2C1
| Supramolecule | Name: Pentamer complex of gH/gL-gp42 with fab 2C1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) |
-Macromolecule #1: Soluble gp42
| Macromolecule | Name: Soluble gp42 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) |
| Molecular weight | Theoretical: 22.398324 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGRVAAAAIT WVPKPNVEVW PVDPPPPVNF NKTAEQEYGD KEVKLPHWTP TLHTFQVPQN YTKANCTYCN TREYTFSYKG CCFYFTKKK HTWNGCFQAC AELYPCTYFY GPTPDILPVV TRNLNAIESL WVGVYRVGEG NWTSLDGGTF KVYQIFGSHC T YVSKFSTV ...String: GGRVAAAAIT WVPKPNVEVW PVDPPPPVNF NKTAEQEYGD KEVKLPHWTP TLHTFQVPQN YTKANCTYCN TREYTFSYKG CCFYFTKKK HTWNGCFQAC AELYPCTYFY GPTPDILPVV TRNLNAIESL WVGVYRVGEG NWTSLDGGTF KVYQIFGSHC T YVSKFSTV PVSHHECSFL KPCLCVSQRS NSHHHHHH UniProtKB: Glycoprotein 42 |
-Macromolecule #2: 2C1 heavy chain
| Macromolecule | Name: 2C1 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.978212 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLVQSGAEVK KPGSSVKVSC RASGGTFDTY AISWVRQAPG HGLEWMGGII PVSGTANYAQ KFQGRVTITA DESTGTAYMD LSSLRSEDT AVYYCARVPD YGTNTPFDYW GQ |
-Macromolecule #3: 2C1 light chain
| Macromolecule | Name: 2C1 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.849269 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIRLTQSPSS LPASVGDRVT ITCRASQDIA TYLAWYQQKP GRAPNLLIYA TSTLQSGVPP RFSGSRSGTD FTLTISSLQP EDFATYYCQ QLRTYPITFG QGTRLEIK |
-Macromolecule #4: Envelope glycoprotein H
| Macromolecule | Name: Envelope glycoprotein H / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) |
| Molecular weight | Theoretical: 72.600352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LSEVKLHLDI EGHASHYTIP WTELMAKVPG LSPEALWREA NVTEDLASML NRYKLIYKTS GTLGIALAEP VDIPAVSEGS MQVDASKVH PGVISGLNSP ACMLSAPLEK QLFYYIGTML PNTRPHSYVF YQLRCHLSYV ALSINGDKFQ YTGAMTSKFL M GTYKRVTE ...String: LSEVKLHLDI EGHASHYTIP WTELMAKVPG LSPEALWREA NVTEDLASML NRYKLIYKTS GTLGIALAEP VDIPAVSEGS MQVDASKVH PGVISGLNSP ACMLSAPLEK QLFYYIGTML PNTRPHSYVF YQLRCHLSYV ALSINGDKFQ YTGAMTSKFL M GTYKRVTE KGDEHVLSLV FGKTKDLPDL RGPFSYPSLT SAQSGDYSLV IVTTFVHYAN FHNYFVPNLK DMFSRAVTMT AA SYARYVL QKLVLLEMKG GCREPELDTE TLTTMFEVSV AFFKVGHAVG ETGNGCVDLR WLAKSFFELT VLKDIIGICY GAT VKGMQS YGLERLAAML MATVKMEELG HLTTEKQEYA LRLATVGYPK AGVYSGLIGG ATSVLLSAYN RHPLFQPLHT VMRE TLFIG SHVVLRELRL NVTTQGPNLA LYQLLSTALC SALEIGEVLR GLALGTESGL FSPCYLSLRF DLTRDKLLSM APQEA TLDQ AAVSNAVDGF LGRLSLERED RDAWHLPAYK CVDRLDKVLM IIPLINVTFI ISSDREVRGS ALYEASTTYL SSSLFL SPV IMNKCSQGAV AGEPRQIPKI QNFTRTQKSC IFCGFALLSY DEKEGLETTT YITSQEVQNS ILSSNYFDFD NLHVHYL LL TTNGTVMEIA GLY UniProtKB: BXLF2 |
-Macromolecule #5: Envelope glycoprotein L
| Macromolecule | Name: Envelope glycoprotein L / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Epstein-Barr virus (strain GD1) (Epstein-Barr virus) Epstein-Barr virus (strain GD1) (Epstein-Barr virus) |
| Molecular weight | Theoretical: 11.759323 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PCCHVTQLRA QHLLALENIS DIYLVSNQTC DGFSLASLNS PKNGSNQLVI SRCANGLNVV SFFISILKRS SSALTGHLRE LLTTLETLY GSFSVEDLFG ANLNRYAW UniProtKB: BKRF2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)