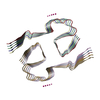
Entry Database : EMDB / ID : EMD-37198Title The cryo-EM structure of type1 amyloid beta 42 fibril in AD2 patient. Tissue : amyloid betaProtein or peptide : Amyloid-beta precursor protein / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 3.0 Å Zhao QY / Tao YQ / Liu C / Li D Funding support 1 items Organization Grant number Country Not funded
Journal : To Be Published Title : The cryo-EM structure of type1 amyloid beta 42 fibril in AD2 patient.Authors : Zhao QY / Tao YQ / Liu C / Li D History Deposition Aug 15, 2023 - Header (metadata) release Aug 21, 2024 - Map release Aug 21, 2024 - Update Jun 25, 2025 - Current status Jun 25, 2025 Processing site : PDBj / Status : Released
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information
 Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Downloads & links
Downloads & links emd_37198.map.gz
emd_37198.map.gz EMDB map data format
EMDB map data format emd-37198-v30.xml
emd-37198-v30.xml emd-37198.xml
emd-37198.xml EMDB header
EMDB header emd_37198_fsc.xml
emd_37198_fsc.xml FSC data file
FSC data file emd_37198.png
emd_37198.png emd-37198.cif.gz
emd-37198.cif.gz emd_37198_half_map_1.map.gz
emd_37198_half_map_1.map.gz emd_37198_half_map_2.map.gz
emd_37198_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-37198
http://ftp.pdbj.org/pub/emdb/structures/EMD-37198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37198
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37198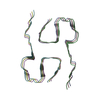
 F&H Search
F&H Search Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
Map Download / File: emd_37198.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_37198.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Sample components
Sample components Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)