+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the wild-typed alpha-galactosidase 5 | |||||||||
 Map data Map data | This map is the wild type of AGAL5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alpha-Galactosidase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationalpha-galactosidase / alpha-galactosidase activity / carbohydrate catabolic process Similarity search - Function | |||||||||
| Biological species |  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) | |||||||||
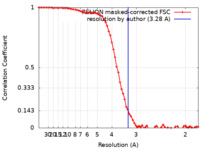
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Li YW / Ru YX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Activity-Based Meta proteomics Drives Discovery and Enzymological Characterization of Novel alpha-galactosidases in the Gut Microbiome Authors: Li YW / Ru YX | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36790.map.gz emd_36790.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36790-v30.xml emd-36790-v30.xml emd-36790.xml emd-36790.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36790_fsc.xml emd_36790_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_36790.png emd_36790.png | 94.3 KB | ||
| Filedesc metadata |  emd-36790.cif.gz emd-36790.cif.gz | 5.5 KB | ||
| Others |  emd_36790_half_map_1.map.gz emd_36790_half_map_1.map.gz emd_36790_half_map_2.map.gz emd_36790_half_map_2.map.gz | 23.2 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36790 http://ftp.pdbj.org/pub/emdb/structures/EMD-36790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36790 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36790 | HTTPS FTP |
-Validation report
| Summary document |  emd_36790_validation.pdf.gz emd_36790_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36790_full_validation.pdf.gz emd_36790_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_36790_validation.xml.gz emd_36790_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_36790_validation.cif.gz emd_36790_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36790 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36790 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36790 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36790 | HTTPS FTP |
-Related structure data
| Related structure data |  8k1aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36790.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36790.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is the wild type of AGAL5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
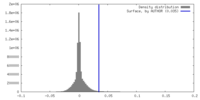
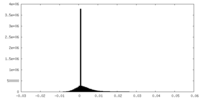
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: This is the half map 2
| File | emd_36790_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
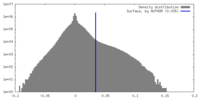
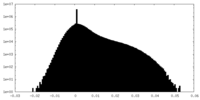
| Density Histograms |
-Half map: This is the half map 1
| File | emd_36790_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetramer complex of alpha-galactosidase 5
| Entire | Name: Tetramer complex of alpha-galactosidase 5 |
|---|---|
| Components |
|
-Supramolecule #1: Tetramer complex of alpha-galactosidase 5
| Supramolecule | Name: Tetramer complex of alpha-galactosidase 5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) |
-Macromolecule #1: Alpha-galactosidase
| Macromolecule | Name: Alpha-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) |
| Molecular weight | Theoretical: 88.017266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAVIFHEKTK EFHIFNREVS YLMRIMENGQ LENLYYGKVI RDKEDFGYLH EEAMRSQMSV CIPEPGILSM QYTRQEYPVY GTGDYRSPA LTVLQENGSR LVDFSYVSHE IYKGKKGIPP LPSTYAESED EAETLEVTLH DQVTDTDLVL TYTIYEDYPV I TRNARFEQ ...String: MAVIFHEKTK EFHIFNREVS YLMRIMENGQ LENLYYGKVI RDKEDFGYLH EEAMRSQMSV CIPEPGILSM QYTRQEYPVY GTGDYRSPA LTVLQENGSR LVDFSYVSHE IYKGKKGIPP LPSTYAESED EAETLEVTLH DQVTDTDLVL TYTIYEDYPV I TRNARFEQ KGEQKIVLER AMSASVEFLD MDYELVQLSG AWSRERYVKN RKLEMGIQSV HSLNGTCGGA EHNPFIALKR PQ TTENQGE VYGFSLVYSG NFLAQAEVST FDMTRVMLGI NPEDFSWELN QGESFQTPEV VMVYSDRGLN KMSQAYHRLY RTR LMRVTW RDKARPILLN NWEATYFDFN EEKILKIAEK AKEAGVELFV LDDGWFGARN DDYRGLGDWY VNLEKLPDGI AGLS RKVEA LGLKFGLWVE LEMVNKDSDL YRAHPDWLIG APDRFESHAR HQHVLDFSRK EVVDYIYKMI AKVLRESSIS YIKWD MNRY MTEPYSRGAD ASQQGKVMHK YILGVYDLYT RLTTEFPEIL FESCASGGAR FDPAMLYFAP QTWTSDDTDA SERTKI QYG TSYVYPVVSM GSHVSAVPNH QMHRMTPIET RANVAYFGTF GYELDLNLLS EAELESVKKQ IAFMKEYREL IQVDGDF YR LLSPFEGNET AWMVVAQDKS RAVAAFYQRM NKVNASWIRF KLQGLDAGTL YEVSCDMAPS ASYDESLAKI YGIQTEEN M VKTYRAYGDE LMQVGIPIDR EDLNKKGGDF ASLLYTLKKV TD UniProtKB: Alpha-galactosidase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



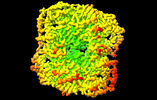
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)