+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the NADP-dependent malic enzyme MaeB | |||||||||
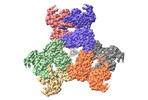
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type III secretion system / inositol phosphate phosphatase / bacteria effector / LIPID BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmalolactic enzyme activity / malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) / malate dehydrogenase (decarboxylating) (NADP+) activity / oxaloacetate decarboxylase activity / malate metabolic process / acyltransferase activity / NAD binding / manganese ion binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.86 Å | |||||||||
 Authors Authors | Jiang WX / Cheng XQ / Wu M / Ma LX / Xing Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of the NADP-dependent malic enzyme in complex with oxaloacetate Authors: Jiang WX / Cheng XQ / Wu M / Ma LX / Xing Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36749.map.gz emd_36749.map.gz | 406.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36749-v30.xml emd-36749-v30.xml emd-36749.xml emd-36749.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36749.png emd_36749.png | 95.9 KB | ||
| Filedesc metadata |  emd-36749.cif.gz emd-36749.cif.gz | 5.6 KB | ||
| Others |  emd_36749_half_map_1.map.gz emd_36749_half_map_1.map.gz emd_36749_half_map_2.map.gz emd_36749_half_map_2.map.gz | 765.5 MB 765.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36749 http://ftp.pdbj.org/pub/emdb/structures/EMD-36749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36749 | HTTPS FTP |
-Validation report
| Summary document |  emd_36749_validation.pdf.gz emd_36749_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36749_full_validation.pdf.gz emd_36749_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_36749_validation.xml.gz emd_36749_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_36749_validation.cif.gz emd_36749_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36749 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36749 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36749 | HTTPS FTP |
-Related structure data
| Related structure data |  8jzoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36749.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36749.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.56667 Å | ||||||||||||||||||||||||||||||||||||
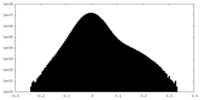
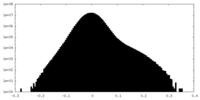
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36749_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
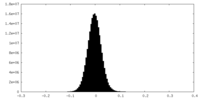
| Density Histograms |
-Half map: #2
| File | emd_36749_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
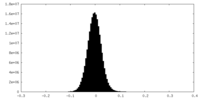
| Density Histograms |
- Sample components
Sample components
-Entire : NADP-dependent malic enzyme
| Entire | Name: NADP-dependent malic enzyme |
|---|---|
| Components |
|
-Supramolecule #1: NADP-dependent malic enzyme
| Supramolecule | Name: NADP-dependent malic enzyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: NADP-dependent malic enzyme
| Macromolecule | Name: NADP-dependent malic enzyme / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.507266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDDQLKQSAL DFHEFPVPGK IQVSPTKPLA TQRDLALAYS PGVAAPCLEI EKDPLKAYKY TARGNLVAVI SNGTAVLGLG NIGALAGKP VMEGKGVLFK KFAGIDVFDI EVDELDPDKF IEVVAALEPT FGGINLEDIK APECFYIEQK LRERMNIPVF H DDQHGTAI ...String: MDDQLKQSAL DFHEFPVPGK IQVSPTKPLA TQRDLALAYS PGVAAPCLEI EKDPLKAYKY TARGNLVAVI SNGTAVLGLG NIGALAGKP VMEGKGVLFK KFAGIDVFDI EVDELDPDKF IEVVAALEPT FGGINLEDIK APECFYIEQK LRERMNIPVF H DDQHGTAI ISTAAILNGL RVVEKNISDV RMVVSGAGAA AIACMNLLVA LGLQKHNIVV CDSKGVIYQG REPNMAETKA AY AVVDDGK RTLDDVIEGA DIFLGCSGPK VLTQEMVKKM ARAPMILALA NPEPEILPPL AKEVRPDAII CTGRSDYPNQ VNN VLCFPF IFRGALDVGA TAINEEMKLA AVRAIAELAH AEQSEVVASA YGDQDLSFGP EYIIPKPFDP RLIVKIAPAV AKAA MESGV ATRPIADFDV YIDKLTEFVY KTNLFMKPIF SQARKAPKRV VLPEGEEARV LHATQELVTL GLAKPILIGR PNVIE MRIQ KLGLQIKAGV DFEIVNNESD PRFKEYWTEY FQIMKRRGVT QEQAQRALIS NPTVIGAIMV QRGEADAMIC GTVGDY HEH FSVVKNVFGY RDGVHTAGAM NALLLPSGNT FIADTYVNDE PDAEELAEIT LMAAETVRRF GIEPRVALLS HSNFGSS DC PSSSKMRQAL ELVRERAPEL MIDGEMHGDA ALVEAIRNDR MPDSSLKGSA NILVMPNMEA ARISYNLLRV SSSEGVTV G PVLMGVAKPV HVLTPIASVR RIVNMVALAV VEAQTQPL UniProtKB: NADP-dependent malic enzyme |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.86 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 108061 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)