[English] 日本語
 Yorodumi
Yorodumi- EMDB-36680: The open structure of the mechanosensitive channel MSL10 in Arabi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The open structure of the mechanosensitive channel MSL10 in Arabidopsis thaliana | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mechanosensitive channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprogrammed cell death in response to reactive oxygen species / leaf senescence / detection of mechanical stimulus / mechanosensitive monoatomic ion channel activity / monoatomic anion transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
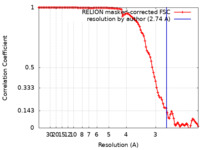
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Sun L / Liu X / Li X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Structural insights into a Plant Mechanosensitive Ion Channel AtMSL10 Authors: Sun L / Liu X / Li X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36680.map.gz emd_36680.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36680-v30.xml emd-36680-v30.xml emd-36680.xml emd-36680.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36680_fsc.xml emd_36680_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36680.png emd_36680.png | 29.6 KB | ||
| Filedesc metadata |  emd-36680.cif.gz emd-36680.cif.gz | 5.3 KB | ||
| Others |  emd_36680_half_map_1.map.gz emd_36680_half_map_1.map.gz emd_36680_half_map_2.map.gz emd_36680_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36680 http://ftp.pdbj.org/pub/emdb/structures/EMD-36680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36680 | HTTPS FTP |
-Validation report
| Summary document |  emd_36680_validation.pdf.gz emd_36680_validation.pdf.gz | 864 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36680_full_validation.pdf.gz emd_36680_full_validation.pdf.gz | 863.6 KB | Display | |
| Data in XML |  emd_36680_validation.xml.gz emd_36680_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_36680_validation.cif.gz emd_36680_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36680 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36680 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36680 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36680 | HTTPS FTP |
-Related structure data
| Related structure data |  8jweMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36680.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36680.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36680_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36680_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heptameric MSL10
| Entire | Name: Heptameric MSL10 |
|---|---|
| Components |
|
-Supramolecule #1: Heptameric MSL10
| Supramolecule | Name: Heptameric MSL10 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mechanosensitive ion channel protein 10
| Macromolecule | Name: Mechanosensitive ion channel protein 10 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 83.138625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEQKSSNGG GGGGDVVINV PVEEASRRSK EMASPESEKG VPFSKSPSPE ISKLVGSPNK PPRAPNQNNV GLTQRKSFAR SVYSKPKSR FVDPSCPVDT SILEEEVREQ LGAGFSFSRA SPNNKSNRSV GSPAPVTPSK VVVEKDEDEE IYKKVKLNRE M RSKISTLA ...String: MAEQKSSNGG GGGGDVVINV PVEEASRRSK EMASPESEKG VPFSKSPSPE ISKLVGSPNK PPRAPNQNNV GLTQRKSFAR SVYSKPKSR FVDPSCPVDT SILEEEVREQ LGAGFSFSRA SPNNKSNRSV GSPAPVTPSK VVVEKDEDEE IYKKVKLNRE M RSKISTLA LIESAFFVVI LSALVASLTI NVLKHHTFWG LEVWKWCVLV MVIFSGMLVT NWFMRLIVFL IETNFLLRRK VL YFVHGLK KSVQVFIWLC LILVAWILLF NHDVKRSPAA TKVLKCITRT LISILTGAFF WLVKTLLLKI LAANFNVNNF FDR IQDSVF HQYVLQTLSG LPLMEEAERV GREPSTGHLS FATVVKKGTV KEKKVIDMGK VHKMKREKVS AWTMRVLMEA VRTS GLSTI SDTLDETAYG EGKEQADREI TSEMEALAAA YHVFRNVAQP FFNYIEEEDL LRFMIKEEVD LVFPLFDGAA ETGRI TRKA FTEWVVKVYT SRRALAHSLN DTKTAVKQLN KLVTAILMVV TVVIWLLLLE VATTKVLLFF STQLVALAFI IGSTCK NLF ESIVFVFVMH PYDVGDRCVV DGVAMLVEEM NLLTTVFLKL NNEKVYYPNA VLATKPISNY FRSPNMGETV EFSISFS TP VSKIAHLKER IAEYLEQNPQ HWAPVHSVVV KEIENMNKLK MALYSDHTIT FQENRERNLR RTELSLAIKR MLEDLHID Y TLLPQDINLT KKN UniProtKB: Mechanosensitive ion channel protein 10 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)