+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Flagellar fibrils from Bacillus amyloliquefaciens | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flagella / motor / fibril / PROTEIN FIBRIL | |||||||||
| Biological species |  | |||||||||
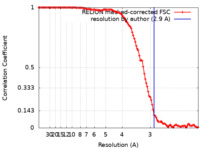
| Method | helical reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Cao Q / Cheng Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Serine peptidase Vpr forms enzymatically active fibrils outside Bacillus bacteria revealed by cryo-EM. Authors: Yijia Cheng / Jianting Han / Meinai Song / Shuqin Zhang / Qin Cao /  Abstract: Bacteria develop a variety of extracellular fibrous structures crucial for their survival, such as flagella and pili. In this study, we use cryo-EM to identify protein fibrils surrounding lab- ...Bacteria develop a variety of extracellular fibrous structures crucial for their survival, such as flagella and pili. In this study, we use cryo-EM to identify protein fibrils surrounding lab-cultured Bacillus amyloiquefaciens and discover an unreported fibril species in addition to the flagellar fibrils. These previously unknown fibrils are composed of Vpr, an extracellular serine peptidase. We find that Vpr assembles into fibrils in an enzymatically active form, potentially representing a strategy of enriching Vpr activities around bacterial cells. Vpr fibrils are also observed under other culture conditions and around other Bacillus bacteria, such as Bacillus subtilis, which may suggest a general mechanism across all Bacillus bacterial groups. Taken together, our study reveals fibrils outside the bacterial cell and sheds light on the physiological role of these extracellular fibrils. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36427.map.gz emd_36427.map.gz | 110.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36427-v30.xml emd-36427-v30.xml emd-36427.xml emd-36427.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36427_fsc.xml emd_36427_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_36427.png emd_36427.png | 85.6 KB | ||
| Filedesc metadata |  emd-36427.cif.gz emd-36427.cif.gz | 4.9 KB | ||
| Others |  emd_36427_half_map_1.map.gz emd_36427_half_map_1.map.gz emd_36427_half_map_2.map.gz emd_36427_half_map_2.map.gz | 97.2 MB 97.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36427 http://ftp.pdbj.org/pub/emdb/structures/EMD-36427 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36427 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36427 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36427.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36427.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36427_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
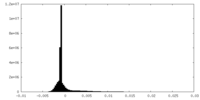
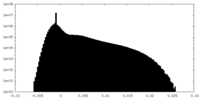
| Density Histograms |
-Half map: #1
| File | emd_36427_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
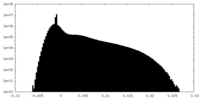
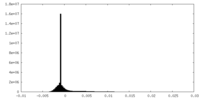
| Density Histograms |
- Sample components
Sample components
-Entire : Bacillus amyloliquefaciens
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus amyloliquefaciens
| Supramolecule | Name: Bacillus amyloliquefaciens / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Flagella
| Macromolecule | Name: Flagella / type: protein_or_peptide / ID: 1 / Number of copies: 33 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.12884 KDa |
| Sequence | String: MRINHNIAAL NTSRQLNAGS NAASKNMEKL SSGLRINRAG DDAAGLAISE KMRSQIRGLD MASKNAQDGI SLIQTSEGAL NETHSILQR MSELATQAAN DTNTDSDRSE LQKEMDQLAS EVTRISTDTE FNTKKLLDGT AQNLTFQIGA NEGQTMSLSI N KMDSESLK ...String: MRINHNIAAL NTSRQLNAGS NAASKNMEKL SSGLRINRAG DDAAGLAISE KMRSQIRGLD MASKNAQDGI SLIQTSEGAL NETHSILQR MSELATQAAN DTNTDSDRSE LQKEMDQLAS EVTRISTDTE FNTKKLLDGT AQNLTFQIGA NEGQTMSLSI N KMDSESLK VGTTYTANDD GSKLVTADGK EATLVTDGSK GPNGYYDDAD KLVYQADSAL AKDTKVTKGI DISSSAKAAS SA LTTIKTA IDTVSSERAK LGAVQNRLEH TINNLGTSSE NLTSAESRIR DVDMASEMME YTKNNILTQA SQAMLAQANQ QPQ QVLQLL KG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.6000000000000005 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)