[English] 日本語
 Yorodumi
Yorodumi- EMDB-35865: Structure of the Mumps Virus L Protein (state2) Bound by Phosphop... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Mumps Virus L Protein (state2) Bound by Phosphoprotein Tetramer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mumps virus polymerase complex / RNA-dependent RNA synthesis / Large protein / phosphoprotein. / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / hydrolase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / ATP binding Similarity search - Function | |||||||||
| Biological species |   Mumps virus strain Jeryl Lynn Mumps virus strain Jeryl Lynn | |||||||||
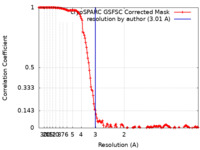
| Method | single particle reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Li TH / Shen QT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural insights into mumps virus polymerase complex for coordinating replication and transcription Authors: Li TL / Shen QT | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35865.map.gz emd_35865.map.gz | 190.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35865-v30.xml emd-35865-v30.xml emd-35865.xml emd-35865.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35865_fsc.xml emd_35865_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_35865.png emd_35865.png | 31.4 KB | ||
| Filedesc metadata |  emd-35865.cif.gz emd-35865.cif.gz | 7.2 KB | ||
| Others |  emd_35865_half_map_1.map.gz emd_35865_half_map_1.map.gz emd_35865_half_map_2.map.gz emd_35865_half_map_2.map.gz | 301.1 MB 301.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35865 http://ftp.pdbj.org/pub/emdb/structures/EMD-35865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35865 | HTTPS FTP |
-Related structure data
| Related structure data |  8izmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35865.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35865.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.53 Å | ||||||||||||||||||||||||||||||||||||
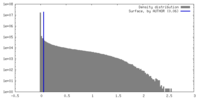
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35865_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
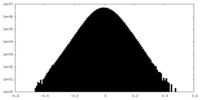
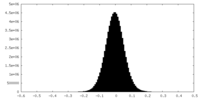
| Density Histograms |
-Half map: #2
| File | emd_35865_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
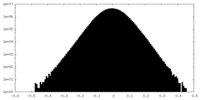
| Density Histograms |
- Sample components
Sample components
-Entire : The MuV polymerase complex of RNA-directed RNA polymerase L with ...
| Entire | Name: The MuV polymerase complex of RNA-directed RNA polymerase L with tetrameric phosphoproteins |
|---|---|
| Components |
|
-Supramolecule #1: The MuV polymerase complex of RNA-directed RNA polymerase L with ...
| Supramolecule | Name: The MuV polymerase complex of RNA-directed RNA polymerase L with tetrameric phosphoproteins type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The MuV polymerase complex expressed in Sf9 cells and purified by affinity chromatography and size-exlusive chromatography sequentially. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mumps virus strain Jeryl Lynn / Strain: Jeryl-Lynn Mumps virus strain Jeryl Lynn / Strain: Jeryl-Lynn |
| Molecular weight | Theoretical: 41.651066 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDQFIKQDET GDLIETGMNV ANHFLSTPIQ GTNSLSKASI LPGVAPVLIG NPEQKNIQHP TASHQGSKTK GRGSGVRSII VSPSEAGNG GTQIPEPLFA QTGQGGIVTT VYQDPTIQPT GSYRSVELAK IGKERMINRF VEKPRTSTPV TEFKRGGPGA A AQGQTIQE ...String: MDQFIKQDET GDLIETGMNV ANHFLSTPIQ GTNSLSKASI LPGVAPVLIG NPEQKNIQHP TASHQGSKTK GRGSGVRSII VSPSEAGNG GTQIPEPLFA QTGQGGIVTT VYQDPTIQPT GSYRSVELAK IGKERMINRF VEKPRTSTPV TEFKRGGPGA A AQGQTIQE EGIDGNGASA GSKERSGSLS GATLYAHLSL PQQDSTPANV GIAPQSAISA NEIMDLLRGM DARLQHLEQK VD KVLAQGS MVTQIKNELS TVKTTLATIE GMMATVKIMD PGNPTGVPVD ELRRSFSDHV TIVSGPGDVS FSSSEKPTLY LDE LARPVS KPRPAKQTKS QPVKDLAGQK VMITKMITDC VANPQMKQAF EQRLAKASTE DALNDIKRDI IRSAI UniProtKB: Phosphoprotein |
-Macromolecule #2: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Mumps virus strain Jeryl Lynn / Strain: Jeryl-Lynn Mumps virus strain Jeryl Lynn / Strain: Jeryl-Lynn |
| Molecular weight | Theoretical: 256.833094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGLNEILLP EVHLNSPIVR YKLFYYILHG QLPNDLEPDD LGPLANQNWK AIRAEESQVH ARLKQIRVEL IARIPSLRWT RSQREIAIL IWPRILPILQ AYDLRQSMQL PTVWEKLTQS TVNLISDGLE RVVLHISNQL TGKPNLFTRS RAGQDTKDYS I PSTRELSQ ...String: MAGLNEILLP EVHLNSPIVR YKLFYYILHG QLPNDLEPDD LGPLANQNWK AIRAEESQVH ARLKQIRVEL IARIPSLRWT RSQREIAIL IWPRILPILQ AYDLRQSMQL PTVWEKLTQS TVNLISDGLE RVVLHISNQL TGKPNLFTRS RAGQDTKDYS I PSTRELSQ IWFNNEWSGS VKTWLMIKYR MRQLITNQKT GELTDLVTIV DTRSTLCIIT PELVALYSSE HKALTYLTFE MV LMVTDML EGRLNVSSLC TASHYLSPLK KRIEVLLTLV DDLALLMGDK VYGIVSSLES FVYAQLQYGD PVIDIKGTFY GFI CNEILD LLTEDNIFTE EEANKVLLDL TSQFDNLSPD LTAELLCIMR LWGHPTLTAS QAASKVRESM CAPKVLDFQT IMKT LAFFH AILINGYRRS HNGIWPPTTL HGNAPKSLIE MRHDNSELKY EYVLKNWKSI SMLRIHKCFD ASPDEDLSIF MKDKA ISCP RQDWMGVFRR SLIKQRYRDA NRPLPQPFNR RLLLNFLEDD RFDPIKELEY VTSGEYLRDP EFCASYSLKE KEIKAT GRI FAKMTKRMRS CQVIAESLLA NHAGKLMREN GVVLDQLKLT KSLLTMNQIG IISEHSRRST ADNMTLAHSG SNKHRIN NS QFKKNKDNKH EMPDDGFEIA ACFLTTDLTK YCLNWRYQVI IPFARTLNSM YGIPHLFEWI HLRLMRSTLY VGDPFNPP S DPTQLDLDTA LNDDIFIVSP RGGIEGLCQK LWTMISISTI ILSATEANTR VMSMVQGDNQ AIAITTRVVR SLSHSEKKE QAYKASKLFF ERLRANNHGI GHHLKEQETI LSSDFFIYSK RVFYKGRILT QALKNVSKMC LTADILGDCS QASCSNLATT VMRLTENGV EKDLCYFLNA FMTIRQLCYD LVFPQTKSLS QDITNAYLNH PILISRLCLL PSQLGGLNFL SCSRLFNRNI G DPLVSAIA DVKRLIKAGC LDIWVLYNIL GRRPGKGKWS TLAADPYTLN IDYLVPSTTF LKKHAQYTLM ERSVNPMLRG VF SENAAEE EEELAQYLLD REVVMPRVAH VILAQSSCGR RKQIQGYLDS TRTIIRYSLE VRPLSAKKLN TVIEYNLLYL SYN LEIIEK PNIVQPFLNA INVDTCSIDI ARSLRKLSWA TLLNGRPIEG LETPDPIELV HGCLIIGSDE CEHCSSGDDK FTWF FLPKG IRLDDDPASN PPIRVPYIGS KTDERRVASM AYIKGASVSL KSALRLAGVY IWAFGDTEES WQDAYELAST RVNLT LEQL QSLTPLPTSA NLVHRLDDGT TQLKFTPASS YAFSSFVHIS NDCQILEIDD QVTDSNLIYQ QVMITGLALI ETWNNP PIN FSVYETTLHL HTGSSCCIRP VESCVVNPPL LPVPLINVPQ MNKFVYDPEP LSLLEMEKIE DIAYQTRIGG LDQIPLL EK IPLLAHLTAK QMVNSITGLD EATSIMNDAV VQADYTSNWI SECCYTYIDS VFVYSGWALL LELSYQMYYL RIQGIQGI L DYVYMTLRRI PGMAITGISS TISHPRILRR CINLDVIAPI NSPHIASLDY TKLSIDAVMW GTKQVLTNIS QGIDYEIVV PSESQLTLSD RVLNLVARKL SLLAIIWANY NYPPKVKGMS PEDKCQALTT HLLQTVEYVE YIQIEKTNIR RMIIEPKLTA YPSNLFYLS RKLLNAIRDS EEGQFLIASY YNSFGYLEPI LMESKIFNLS SSESASLTEF DFILNLELSD ASLEKYSLPS L LMTAENMD NPFPQPPLHH VLRPLGLSST SWYKTISVLN YISHMKISDG AHLYLAEGSG ASMSLIETFL PGETIWYNSL FN SGENPPQ RNFAPLPTQF IESVPYRLIQ AGIAAGNGIV QSFYPLWNGN SDITDLSTKT SVEYIIHKVG ADTCALVHVD LEG VPGSMN SMLERAQVHA LLITVTVLKP GGLLILKASW EPFNRFSFLL TVLWQFFSTI RILRSSYSDP NNHEVYIIAT LAVD PTTSS FTTALNRART LNEQGFSLIP PELVSEYWRK RVEQGQIIQD CIDKVISECV RDQYLADNNI ILQAGGTPST RKWLD LPDY SSFNELQSEM ARLITIHLKE VIEILKGQAS DHDTLLFTSY NVGPLGKINT ILRLIVERIL MYTVRNWCIL PTQTRL TLR QSIELGEFRL RDVITPMEIL KLSPNRKYLK SALNQSTFNH LMGETSDILL NRAYQKRIWK AIGCVIYCFG LLTPDVE GS ERIDVDNDIP DYDIHGDII UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.75 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)