[English] 日本語
 Yorodumi
Yorodumi- EMDB-35432: C2 reconstruction of the concave tetramer in the cube-like assemb... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C2 reconstruction of the concave tetramer in the cube-like assembly of 201Phi2-1 gp105 | |||||||||
 Map data Map data | The cryo-EM map of the phage-encoded nucleus-like shell protein gp105 concave tetramer reconstructed with C2 symmetry imposed. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | jumbo phage 201phi2-1 / gp105 / nucleus-like structure. / STRUCTURAL PROTEIN | |||||||||
| Function / homology | host cell cytoplasm / Chimallin Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) | |||||||||
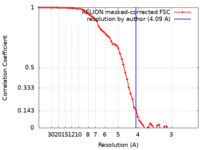
| Method | single particle reconstruction / cryo EM / Resolution: 4.09 Å | |||||||||
 Authors Authors | Liu Z / Xiang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Front Microbiol / Year: 2023 Journal: Front Microbiol / Year: 2023Title: Structural studies of the nucleus-like assembly of jumbo bacteriophage 201φ2-1. Authors: Zhe Liu / Ye Xiang /  Abstract: The jumbo phages encode proteins that assemble to form a nucleus-like compartment in infected cells. Here we report the cryo-EM structure and biochemistry characterization of gp105, a protein that is ...The jumbo phages encode proteins that assemble to form a nucleus-like compartment in infected cells. Here we report the cryo-EM structure and biochemistry characterization of gp105, a protein that is encoded by the jumbo phage 201φ2-1 and is involved in the formation of the nucleus-like compartment in phage 201φ2-1 infected . We found that, although most gp105 molecules are in the monomeric state in solution, a small portion of gp105 assemble to form large sheet-like assemblies and small cube-like particles. Reconstruction of the cube-like particles showed that the particle consists of six flat head-to-tail tetramers arranged into an octahedral cube. The four molecules at the contact interface of two head-to-tail tetramers are 2-fold symmetry-related and constitute a concave tetramer. Further reconstructions without applying symmetry showed that molecules in the particles around the distal ends of a 3-fold axis are highly dynamic and have the tendency to open up the assembly. Local classifications and refinements of the concave tetramers in the cube-like particle resulted in a map of the concave tetramer at a resolution of 4.09 Å. Structural analysis of the concave tetramer indicates that the N and C terminal fragments of gp105 are important for mediating the intermolecular interactions, which was further confirmed by mutagenesis studies. Biochemistry assays showed that, in solution, the cube-like particles of gp105 are liable to either disassemble to form the monomers or recruit more molecules to form the high molecular weight lattice-like assembly. We also found that monomeric gp105s can self-assemble to form large sheet-like assemblies , and the assembly of gp105 is a reversible dynamic process and temperature-dependent. Taken together, our results revealed the dynamic assembly of gp105, which helps to understand the development and function of the nucleus-like compartment assembled by phage-encoded proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35432.map.gz emd_35432.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35432-v30.xml emd-35432-v30.xml emd-35432.xml emd-35432.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35432_fsc.xml emd_35432_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_35432.png emd_35432.png | 156.6 KB | ||
| Filedesc metadata |  emd-35432.cif.gz emd-35432.cif.gz | 5.5 KB | ||
| Others |  emd_35432_half_map_1.map.gz emd_35432_half_map_1.map.gz emd_35432_half_map_2.map.gz emd_35432_half_map_2.map.gz | 16.1 MB 16.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35432 http://ftp.pdbj.org/pub/emdb/structures/EMD-35432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35432 | HTTPS FTP |
-Related structure data
| Related structure data |  8iggMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35432.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35432.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
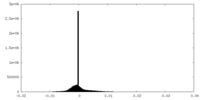
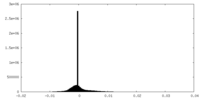
| Annotation | The cryo-EM map of the phage-encoded nucleus-like shell protein gp105 concave tetramer reconstructed with C2 symmetry imposed. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: The cryo-EM half map of the phage-encoded nucleus-like...
| File | emd_35432_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
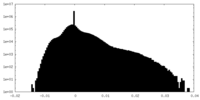
| Annotation | The cryo-EM half map of the phage-encoded nucleus-like shell protein gp105 concave tetramer reconstructed with C2 symmetry imposed. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The cryo-EM half map of the phage-encoded nucleus-like...
| File | emd_35432_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
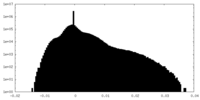
| Annotation | The cryo-EM half map of the phage-encoded nucleus-like shell protein gp105 concave tetramer reconstructed with C2 symmetry imposed. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 201Phi2-1 gp105
| Entire | Name: 201Phi2-1 gp105 |
|---|---|
| Components |
|
-Supramolecule #1: 201Phi2-1 gp105
| Supramolecule | Name: 201Phi2-1 gp105 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) |
-Macromolecule #1: Chimallin
| Macromolecule | Name: Chimallin / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) |
| Molecular weight | Theoretical: 70.654516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIRDTATNTT QTQAAPQQAP AQQFTQAPQE KPMQSTQSQP TPSYAGTGGI NSQFTRSGNV QGGDARASEA LTVFTRLKEQ AVAQQDLAD DFSILRFDRD QHQVGWSSLV IAKQISLNGQ PVIAVRPLIL PNNSIELPKR KTNIVNGMQT DVIESDIDVG T VFSAQYFN ...String: MIRDTATNTT QTQAAPQQAP AQQFTQAPQE KPMQSTQSQP TPSYAGTGGI NSQFTRSGNV QGGDARASEA LTVFTRLKEQ AVAQQDLAD DFSILRFDRD QHQVGWSSLV IAKQISLNGQ PVIAVRPLIL PNNSIELPKR KTNIVNGMQT DVIESDIDVG T VFSAQYFN RLSTYVQNTL GKPGAKVVLA GPFPIPADLV LKDSELQLRN LLIKSVNACD DILALHSGER PFTIAGLKGQ QG ETLAAKV DIRTQPLHDT VGNPIRADIV VTTQRVRRNG QQENEFYETD VKLNQVAMFT NLERTPQAQA QTLFPNQQQV ATP APWVAS VVITDVRNAD GIQANTPEMY WFALSNAFRS THGHAWARPF LPMTGVAKDM KDIGALGWMS ALRNRIDTKA ANFD DAQFG QLMLSQVQPN PVFQIDLNRM GETAQMDSLQ LDAAGGPNAQ KAAATIIRQI NNLGGGGFER FFDHTTQPIL ERTGQ VIDL GNWFDGDEKR DRRDLDNLAA LNAAEGNENE FWGFYGAQLN PNLHPDLRNR QSRNYDRQYL GSTVTYTGKA ERCTYN AKF IEALDRYLAE AGLQITMDNT SVLNSGQRFM GNSVIGNNMV SGQAQVHSAY AGTQGFNTQY QTGPSSFYAL EHHHHHH UniProtKB: Chimallin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)