+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP13A2 in the E1-ATP state | |||||||||
 Map data Map data | Cryo-EM structure of ATP13A2 in the E1-ATP state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM structure of ATP13A2 in the E1-ATP state / Membran protein / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolyamine transmembrane transporter activity / polyamine transmembrane transport / spermine transmembrane transport / peptidyl-aspartic acid autophosphorylation / regulation of ubiquitin-specific protease activity / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / P-type ion transporter activity / regulation of chaperone-mediated autophagy ...polyamine transmembrane transporter activity / polyamine transmembrane transport / spermine transmembrane transport / peptidyl-aspartic acid autophosphorylation / regulation of ubiquitin-specific protease activity / ABC-type polyamine transporter activity / regulation of autophagosome size / extracellular exosome biogenesis / P-type ion transporter activity / regulation of chaperone-mediated autophagy / negative regulation of lysosomal protein catabolic process / regulation of autophagy of mitochondrion / regulation of lysosomal protein catabolic process / intracellular monoatomic cation homeostasis / autophagosome-lysosome fusion / autophagosome organization / protein localization to lysosome / phosphatidic acid binding / positive regulation of exosomal secretion / multivesicular body membrane / ATPase-coupled monoatomic cation transmembrane transporter activity / intracellular zinc ion homeostasis / regulation of protein localization to nucleus / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / cupric ion binding / regulation of mitochondrion organization / regulation of endopeptidase activity / lysosomal transport / phosphatidylinositol-3,5-bisphosphate binding / cellular response to zinc ion / regulation of intracellular protein transport / lipid homeostasis / Ion transport by P-type ATPases / autophagosome membrane / cellular response to manganese ion / regulation of macroautophagy / regulation of neuron apoptotic process / transport vesicle / autophagosome / multivesicular body / lysosomal lumen / positive regulation of protein secretion / transmembrane transport / autophagy / intracellular calcium ion homeostasis / late endosome / late endosome membrane / manganese ion binding / cellular response to oxidative stress / monoatomic ion transmembrane transport / intracellular iron ion homeostasis / vesicle / protein autophosphorylation / lysosome / neuron projection / lysosomal membrane / neuronal cell body / positive regulation of gene expression / ATP hydrolysis activity / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Liu ZM / Mu JQ / Xue CY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Conformational cycle of human polyamine transporter ATP13A2. Authors: Jianqiang Mu / Chenyang Xue / Lei Fu / Zongjun Yu / Minhan Nie / Mengqi Wu / Xinmeng Chen / Kun Liu / Ruiqian Bu / Ying Huang / Baisheng Yang / Jianming Han / Qianru Jiang / Kevin C Chan / ...Authors: Jianqiang Mu / Chenyang Xue / Lei Fu / Zongjun Yu / Minhan Nie / Mengqi Wu / Xinmeng Chen / Kun Liu / Ruiqian Bu / Ying Huang / Baisheng Yang / Jianming Han / Qianru Jiang / Kevin C Chan / Ruhong Zhou / Huilin Li / Ancheng Huang / Yong Wang / Zhongmin Liu /  Abstract: Dysregulation of polyamine homeostasis strongly associates with human diseases. ATP13A2, which is mutated in juvenile-onset Parkinson's disease and autosomal recessive spastic paraplegia 78, is a ...Dysregulation of polyamine homeostasis strongly associates with human diseases. ATP13A2, which is mutated in juvenile-onset Parkinson's disease and autosomal recessive spastic paraplegia 78, is a transporter with a critical role in balancing the polyamine concentration between the lysosome and the cytosol. Here, to better understand human ATP13A2-mediated polyamine transport, we use single-particle cryo-electron microscopy to solve high-resolution structures of human ATP13A2 in six intermediate states, including the putative E2 structure for the P5 subfamily of the P-type ATPases. These structures comprise a nearly complete conformational cycle spanning the polyamine transport process and capture multiple substrate binding sites distributed along the transmembrane regions, suggesting a potential polyamine transport pathway. Integration of high-resolution structures, biochemical assays, and molecular dynamics simulations allows us to obtain a better understanding of the structural basis of how hATP13A2 transports polyamines, providing a mechanistic framework for ATP13A2-related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35384.map.gz emd_35384.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35384-v30.xml emd-35384-v30.xml emd-35384.xml emd-35384.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35384.png emd_35384.png | 29.4 KB | ||
| Filedesc metadata |  emd-35384.cif.gz emd-35384.cif.gz | 6.1 KB | ||
| Others |  emd_35384_half_map_1.map.gz emd_35384_half_map_1.map.gz emd_35384_half_map_2.map.gz emd_35384_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35384 http://ftp.pdbj.org/pub/emdb/structures/EMD-35384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35384 | HTTPS FTP |
-Validation report
| Summary document |  emd_35384_validation.pdf.gz emd_35384_validation.pdf.gz | 748.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35384_full_validation.pdf.gz emd_35384_full_validation.pdf.gz | 748.1 KB | Display | |
| Data in XML |  emd_35384_validation.xml.gz emd_35384_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_35384_validation.cif.gz emd_35384_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35384 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35384 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35384 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35384 | HTTPS FTP |
-Related structure data
| Related structure data |  8iekMC  8ielC  8iemC  8ienC  8ieoC  8ierC  8iesC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35384.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35384.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of ATP13A2 in the E1-ATP state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.095 Å | ||||||||||||||||||||||||||||||||||||
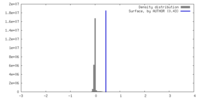
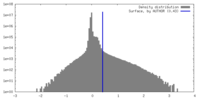
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: The half B map of ATP13A2 in the E1-ATP state
| File | emd_35384_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half_B map of ATP13A2 in the E1-ATP state | ||||||||||||
| Projections & Slices |
| ||||||||||||
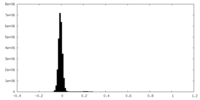
| Density Histograms |
-Half map: The half A map of ATP13A2 in the E1-ATP state
| File | emd_35384_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The half_A map of ATP13A2 in the E1-ATP state | ||||||||||||
| Projections & Slices |
| ||||||||||||
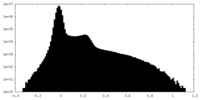
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of ATP13A2 in the E1-ATP state
| Entire | Name: Cryo-EM structure of ATP13A2 in the E1-ATP state |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of ATP13A2 in the E1-ATP state
| Supramolecule | Name: Cryo-EM structure of ATP13A2 in the E1-ATP state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Polyamine-transporting ATPase 13A2
| Macromolecule | Name: Polyamine-transporting ATPase 13A2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 128.914984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSADSSPLVG STPTGYGTLT IGTSIDPLSS SVSSVRLSGY CGSPWRVIGY HVVVWMMAGI PLLLFRWKPL WGVRLRLRPC NLAHAETLV IEIRDKEDSS WQLFTVQVQT EAIGEGSLEP SPQSQAEDGR SQAAVGAVPE GAWKDTAQLH KSEEAVSVGQ K RVLRYYLF ...String: MSADSSPLVG STPTGYGTLT IGTSIDPLSS SVSSVRLSGY CGSPWRVIGY HVVVWMMAGI PLLLFRWKPL WGVRLRLRPC NLAHAETLV IEIRDKEDSS WQLFTVQVQT EAIGEGSLEP SPQSQAEDGR SQAAVGAVPE GAWKDTAQLH KSEEAVSVGQ K RVLRYYLF QGQRYIWIET QQAFYQVSLL DHGRSCDDVH RSRHGLSLQD QMVRKAIYGP NVISIPVKSY PQLLVDEALN PY YGFQAFS IALWLADHYY WYALCIFLIS SISICLSLYK TRKQSQTLRD MVKLSMRVCV CRPGGEEEWV DSSELVPGDC LVL PQEGGL MPCDAALVAG ECMVNESSLT GESIPVLKTA LPEGLGPYCA ETHRRHTLFC GTLILQARAY VGPHVLAVVT RTGF CTAKG GLVSSILHPR PINFKFYKHS MKFVAALSVL ALLGTIYSIF ILYRNRVPLN EIVIRALDLV TVVVPPALPA AMTVC TLYA QSRLRRQGIF CIHPLRINLG GKLQLVCFDK TGTLTEDGLD VMGVVPLKGQ AFLPLVPEPR RLPVGPLLRA LATCHA LSR LQDTPVGDPM DLKMVESTGW VLEEEPAADS AFGTQVLAVM RPPLWEPQLQ AMEEPPVPVS VLHRFPFSSA LQRMSVV VA WPGATQPEAY VKGSPELVAG LCNPETVPTD FAQMLQSYTA AGYRVVALAS KPLPTVPSLE AAQQLTRDTV EGDLSLLG L LVMRNLLKPQ TTPVIQALRR TRIRAVMVTG DNLQTAVTVA RGCGMVAPQE HLIIVHATHP ERGQPASLEF LPMESPTAV NGVKDPDQAA SYTVEPDPRS RHLALSGPTF GIIVKHFPKL LPKVLVQGTV FARMAPEQKT ELVCELQKLQ YCVGMCGDGA NDCGALKAA DVGISLSQAE ASVVSPFTSS MASIECVPMV IREGRCSLDT SFSVFKYMAL YSLTQFISVL ILYTINTNLG D LQFLAIDL VITTTVAVLM SRTGPALVLG RVRPPGALLS VPVLSSLLLQ MVLVTGVQLG GYFLTLAQPW FVPLNRTVAA PD NLPNYEN TVVFSLSSFQ YLILAAAVSK GAPFRRPLYT NVPFLVALAL LSSVLVGLVL VPGLLQGPLA LRNITDTGFK LLL LGLVTL NFVGAFMLES VLDQCLPACL RRLRPKRASK KRFKQLEREL AEQPWPPLPA GPLR UniProtKB: Polyamine-transporting ATPase 13A2 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 325325 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)