[English] 日本語
 Yorodumi
Yorodumi- EMDB-35306: Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ceramide / phosphorylation / TRANSFERASE-INHIBITOR COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of sphingolipid biosynthetic process / positive regulation of sphingolipid biosynthetic process / 3-keto-sphinganine metabolic process / multidimensional cell growth / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / regulation of programmed cell death / ceramide metabolic process ...negative regulation of sphingolipid biosynthetic process / positive regulation of sphingolipid biosynthetic process / 3-keto-sphinganine metabolic process / multidimensional cell growth / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / regulation of programmed cell death / ceramide metabolic process / sphingosine biosynthetic process / embryo development ending in seed dormancy / sphingolipid biosynthetic process / ceramide biosynthetic process / response to unfolded protein / Neutrophil degranulation / enzyme activator activity / pyridoxal phosphate binding / endoplasmic reticulum membrane / endoplasmic reticulum / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Xie T / Gong X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide. Authors: Tian Xie / Feitong Dong / Gongshe Han / Xinyue Wu / Peng Liu / Zike Zhang / Jianlong Zhong / Somashekarappa Niranjanakumari / Kenneth Gable / Sita D Gupta / Wenchen Liu / Peter J Harrison / ...Authors: Tian Xie / Feitong Dong / Gongshe Han / Xinyue Wu / Peng Liu / Zike Zhang / Jianlong Zhong / Somashekarappa Niranjanakumari / Kenneth Gable / Sita D Gupta / Wenchen Liu / Peter J Harrison / Dominic J Campopiano / Teresa M Dunn / Xin Gong /    Abstract: The homeostatic regulation of serine palmitoyltransferase (SPT) activity in yeast involves N-terminal phosphorylation of Orm proteins, while higher eukaryotes lack these phosphorylation sites. ...The homeostatic regulation of serine palmitoyltransferase (SPT) activity in yeast involves N-terminal phosphorylation of Orm proteins, while higher eukaryotes lack these phosphorylation sites. Although recent studies have indicated a conserved ceramide-mediated feedback inhibition of the SPT-ORM/ORMDL complex in higher eukaryotes, its conservation and relationship with phosphorylation regulation in yeast remain unclear. Here, we determine the structure of the yeast SPT-Orm2 complex in a dephosphomimetic state and identify an evolutionarily conserved ceramide-sensing site. Ceramide stabilizes the dephosphomimetic Orm2 in an inhibitory conformation, facilitated by an intramolecular β-sheet between the N- and C-terminal segments of Orm2. Moreover, we find that a phosphomimetic mutant of Orm2, positioned adjacent to its intramolecular β-sheet, destabilizes the inhibitory conformation of Orm2. Taken together, our findings suggest that both Orm dephosphorylation and ceramide binding are crucial for suppressing SPT activity in yeast. This highlights a distinctive regulatory mechanism in yeast involving the collaborative actions of phosphorylation and ceramide. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35306.map.gz emd_35306.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35306-v30.xml emd-35306-v30.xml emd-35306.xml emd-35306.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35306.png emd_35306.png | 64.8 KB | ||
| Filedesc metadata |  emd-35306.cif.gz emd-35306.cif.gz | 7.5 KB | ||
| Others |  emd_35306_half_map_1.map.gz emd_35306_half_map_1.map.gz emd_35306_half_map_2.map.gz emd_35306_half_map_2.map.gz | 48.4 MB 48.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35306 http://ftp.pdbj.org/pub/emdb/structures/EMD-35306 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35306 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35306 | HTTPS FTP |
-Related structure data
| Related structure data |  8iakMC  8iajC  8iamC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35306.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35306.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35306_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
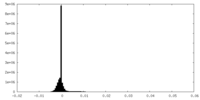
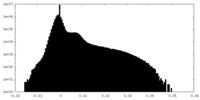
| Density Histograms |
-Half map: #2
| File | emd_35306_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
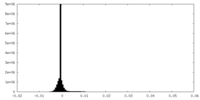
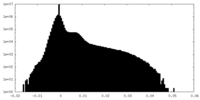
| Density Histograms |
- Sample components
Sample components
-Entire : SPT-ORM2 complex
| Entire | Name: SPT-ORM2 complex |
|---|---|
| Components |
|
-Supramolecule #1: SPT-ORM2 complex
| Supramolecule | Name: SPT-ORM2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Long chain base biosynthesis protein 1,Serine palmitoyltransferase 1
| Macromolecule | Name: Long chain base biosynthesis protein 1,Serine palmitoyltransferase 1 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.354672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASNLVEMFN AALNWVTMIL ESPSARVVLF GVPIRGHFFV EGLLGVVIII LLTRKSYKPP KRPLTEQEID ELCDEWVPEP LVDPSATDE QSWRVAKTPV TMEMPIQNHI TITRNNLQEK YTNVFNLASN NFLQLSATEP VKEVVKTTIK NYGVGACGPA G FYGNQDVH ...String: MASNLVEMFN AALNWVTMIL ESPSARVVLF GVPIRGHFFV EGLLGVVIII LLTRKSYKPP KRPLTEQEID ELCDEWVPEP LVDPSATDE QSWRVAKTPV TMEMPIQNHI TITRNNLQEK YTNVFNLASN NFLQLSATEP VKEVVKTTIK NYGVGACGPA G FYGNQDVH YTLEYDLAQF FGTQGSVLYG QDFCAAPSVL PAFTKRGDVI VADDQVSLPV QNALQLSRST VYYFNHNDMN SL ECLLNEL TEQEKLEKLP AIPRKFIVTE GIFHNSGDLA PLPELTKLKN KYKFRLFVDE TFSIGVLGAT GRGLSEHFNM DRA TAIDIT VGSMATALGS TGGFVLGDSV MCLHQRIGSN AYCFSACLPA YTVTSVSKVL KLMDSNNDAV QTLQKLSKSL HDSF ASDDS LRSYVIVTSS PVSAVLHLQL TPAYRSRKFG YTCEQLFETM SALQKKSQTN KFIEPYEEEE KFLQSIVDHA LINYN VLIT RNTIVLKQET LPIVPSLKIC CNAAMSPEEL KNACESVKQS ILACCQESNK UniProtKB: Long chain base biosynthesis protein 1, Serine palmitoyltransferase 1 |
-Macromolecule #2: Serine palmitoyltransferase 2
| Macromolecule | Name: Serine palmitoyltransferase 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.417832 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSTPANYTRV PLCEPEELPD DIQKENEYGT LDSPGHLYQV KSRHGKPLPE PVVDTPPYYI SLLTYLNYLI LIILGHVHDF LGMTFQKNK HLDLLEHDGL APWFSNFESF YVRRIKMRID DCFSRPTTGV PGRFIRCIDR ISHNINEYFT YSGAVYPCMN L SSYNYLGF ...String: MSTPANYTRV PLCEPEELPD DIQKENEYGT LDSPGHLYQV KSRHGKPLPE PVVDTPPYYI SLLTYLNYLI LIILGHVHDF LGMTFQKNK HLDLLEHDGL APWFSNFESF YVRRIKMRID DCFSRPTTGV PGRFIRCIDR ISHNINEYFT YSGAVYPCMN L SSYNYLGF AQSKGQCTDA ALESVDKYSI QSGGPRAQIG TTDLHIKAEK LVARFIGKED ALVFSMGYGT NANLFNAFLD KK CLVISDE LNHTSIRTGV RLSGAAVRTF KHGDMVGLEK LIREQIVLGQ PKTNRPWKKI LICAEGLFSM EGTLCNLPKL VEL KKKYKC YLFIDEAHSI GAMGPTGRGV CEIFGVDPKD VDILMGTFT(LLP) SFGAAGGYIA ADQWIIDRLR LDLTTVSYSE SMPAPVLAQ TISSLQTISG EICPGQGTER LQRIAFNSRY LRLALQRLGF IVYGVADSPV IPLLLYCPSK MPAFSRMMLQ R RIAVVVVA YPATPLIESR VRFCMSASLT KEDIDYLLRH VSEVGDKLNL KSNSGKSSYD GKRQRWDIEE VIRRTPEDCK DD KYFVN UniProtKB: Serine palmitoyltransferase 2 |
-Macromolecule #3: Protein ORM2
| Macromolecule | Name: Protein ORM2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.787576 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIDRTKNESP AFEESPLTPN VSNLKPFPSQ SNKISTPVTD HRRRRAAAVI SHVEQETFED ENDQQMLPNM AATWVDQRGA WLIHIVVIV LLRLFYSLFG STPKWTWTLT NMTYIIGFYI MFHLVKGTPF DFNGGAYDNL TMWEQINDET LYTPTRKFLL I VPIVLFLI ...String: MIDRTKNESP AFEESPLTPN VSNLKPFPSQ SNKISTPVTD HRRRRAAAVI SHVEQETFED ENDQQMLPNM AATWVDQRGA WLIHIVVIV LLRLFYSLFG STPKWTWTLT NMTYIIGFYI MFHLVKGTPF DFNGGAYDNL TMWEQINDET LYTPTRKFLL I VPIVLFLI SNQYYRNDMT LFLSNLAVTV LIGVVPKLGI THRLRISIPG ITGRAQIS UniProtKB: Protein ORM2 |
-Macromolecule #4: Serine palmitoyltransferase-regulating protein TSC3
| Macromolecule | Name: Serine palmitoyltransferase-regulating protein TSC3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.590233 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTQHKSSMVY IPTTKEAKRR NGKSEGILNT IEEVVEKLYW TYYIHLPFYL MASFDSFFLH VFFLTIFSLS FFGILKYCFL UniProtKB: Serine palmitoyltransferase-regulating protein TSC3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)