[English] 日本語
 Yorodumi
Yorodumi- EMDB-35035: Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA / DNA / RNA BINDING PROTEIN/RNA/DNA / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / defense response to virus / endonuclease activity / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Eubacterium ventriosum ATCC 27560 (bacteria) / synthetic construct (others) Eubacterium ventriosum ATCC 27560 (bacteria) / synthetic construct (others) | |||||||||
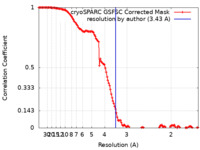
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Tang N / Wu Z / Gao Y / Chen W / Su M / Wang Z / Ji Q | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: ACS Synth Biol / Year: 2024 Journal: ACS Synth Biol / Year: 2024Title: Molecular Basis and Genome Editing Applications of a Compact CRISPR-Cas9 System. Authors: Na Tang / Zhaowei Wu / Yan Gao / Weizhong Chen / Zixiao Wang / Mengjiao Su / Wenxin Ji / Quanjiang Ji /  Abstract: CRISPR-Cas9 systems have been widely harnessed for diverse genome editing applications because of their ease of use and high efficiency. However, the large molecular sizes and strict PAM requirements ...CRISPR-Cas9 systems have been widely harnessed for diverse genome editing applications because of their ease of use and high efficiency. However, the large molecular sizes and strict PAM requirements of commonly used CRISPR-Cas9 systems restrict their broad applications in therapeutics. Here, we report the molecular basis and genome editing applications of a novel compact type II-A CRISPR-Cas9 system (EvCas9) with 1107 residues and distinct 5'-NNGDGN-3' (where D represents A, T, or G) PAM specificity. We determine the cryo-EM structure of EvCas9 in a complex with an sgRNA and a target DNA, revealing the detailed PAM recognition and sgRNA and target DNA association mechanisms. Additionally, we demonstrate the robust genome editing capacity of EvCas9 in bacteria and human cells with superior fidelity compared to SaCas9 and SpCas9, and we engineer it to be efficient base editors by fusing a cytidine or adenosine deaminase. Collectively, our results facilitate further understanding of CRISPR-Cas9 working mechanisms and expand the compact CRISPR-Cas9 toolbox. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35035.map.gz emd_35035.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35035-v30.xml emd-35035-v30.xml emd-35035.xml emd-35035.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35035_fsc.xml emd_35035_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_35035.png emd_35035.png | 101.5 KB | ||
| Filedesc metadata |  emd-35035.cif.gz emd-35035.cif.gz | 6.7 KB | ||
| Others |  emd_35035_half_map_1.map.gz emd_35035_half_map_1.map.gz emd_35035_half_map_2.map.gz emd_35035_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35035 http://ftp.pdbj.org/pub/emdb/structures/EMD-35035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35035 | HTTPS FTP |
-Validation report
| Summary document |  emd_35035_validation.pdf.gz emd_35035_validation.pdf.gz | 777.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35035_full_validation.pdf.gz emd_35035_full_validation.pdf.gz | 776.7 KB | Display | |
| Data in XML |  emd_35035_validation.xml.gz emd_35035_validation.xml.gz | 21.4 KB | Display | |
| Data in CIF |  emd_35035_validation.cif.gz emd_35035_validation.cif.gz | 27.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35035 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35035 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35035 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35035 | HTTPS FTP |
-Related structure data
| Related structure data |  8hudMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35035.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35035.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35035_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
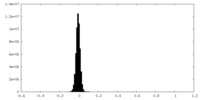
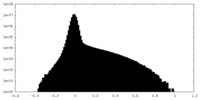
| Density Histograms |
-Half map: #2
| File | emd_35035_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
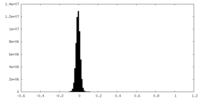
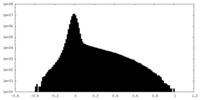
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of EvCas9-sgRNA-dsDNA
| Entire | Name: Ternary complex of EvCas9-sgRNA-dsDNA |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of EvCas9-sgRNA-dsDNA
| Supramolecule | Name: Ternary complex of EvCas9-sgRNA-dsDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Eubacterium ventriosum ATCC 27560 (bacteria) Eubacterium ventriosum ATCC 27560 (bacteria) |
| Molecular weight | Theoretical: 156.5 KDa |
-Macromolecule #1: CRISPR-associated endonuclease Cas9
| Macromolecule | Name: CRISPR-associated endonuclease Cas9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Eubacterium ventriosum ATCC 27560 (bacteria) Eubacterium ventriosum ATCC 27560 (bacteria) |
| Molecular weight | Theoretical: 131.110531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGYTVGLDIG VASVGVAVLD ENDNIVEAVS NIFDEADTSN NKVRRTLREG RRTKRRQKTR IEDFKQLWET SGYIIPHKLH LNIIELRNK GLTELLSLDE LYCVLLSMLK HRGISYLEDA DDGEKGNAYK KGLAFNEKQL KEKMPCEIQL ERMKKYGKYH G EFIIEIND ...String: MGYTVGLDIG VASVGVAVLD ENDNIVEAVS NIFDEADTSN NKVRRTLREG RRTKRRQKTR IEDFKQLWET SGYIIPHKLH LNIIELRNK GLTELLSLDE LYCVLLSMLK HRGISYLEDA DDGEKGNAYK KGLAFNEKQL KEKMPCEIQL ERMKKYGKYH G EFIIEIND EKEYQSNVFT TKAYKKELEK IFETQRCNGN KINTKFIKKY MEIYERKREY YIGPGNEKSR TDYGIYTTRT DE EGNFIDE KNIFGKLIGK CSVYPEEYRA SSASYTAQEF NLLNDLNNLK INNEKLTEFQ KKEIVEIIKD ASSVNMRKII KKV IDEDIE QYSGARIDKK GKEIYHTFEI YRKLKKELKT INVDIDSFTR EELDKTMDIL TLNTERESIV KAFDEQKFVY EENL IKKLI EFRKNNQRLF SGWHSFSYKA MLQLIPVMYK EPKEQMQLLT EMNVFKSKKE KYVNYKYIPE NEVVKEIYNP VVVKS IRTT VKILNALIKK YGYPESVVIE MPRDKNSDDE KEKIDMNQKK NQEEYEKILN KIYDEKGIEI TNKDYKKQKK LVLKLK LWN EQEGLCLYSG KKIAIEDLLN HPEFFEIDHI IPKSISLDDS RSNKVLVYKT ENSIKENDTP YHYLTRINGK WGFDEYK AN VLELRRRGKI DDKKVNNLLC MEDITKIDVV KGFINRNLND TRYASRVVLN EMQSFFESRK YCNTKVKVIR GSLTYQMR Q DLHLKKNREE SYSHHAVDAM LIAFSQKGYE AYRKIQKDCY DFETGEILDK EKWNKYIDDD EFDDILYKER MNEIRKKII EAEEKVKYNY KIDKKCNRGL CNQTIYGTRE KDGKIHKISS YNIYDDKECN SLKKMINSGK GSDLLMYNND PKTYRDMLKI LETYSSEKN PFVAYNKETG DYFRKYSKNH NGPKVEKVKY YSGQINSCID ISHKYGHAKN SKKVVLVSLN PYRTDVYYDN D TGKYYLVG VKYNHIKCVG NKYVIDSETY NELLRKEGVL NSDENLEDLN SKNITYKFSL YKNDIIQYEK GGEYYTERFL SR IKEQKNL IETKPINKPN FQRKNKKGEW ENTRNQIALA KTKYVGKLVT DVLGNCYIVN MEKFSLVVDK UniProtKB: CRISPR-associated endonuclease Cas9 |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.97624 KDa |
| Sequence | String: GGUAAUCGCU CUCCUCCGGC GAUUUUAGUA CCUGAGAAAU CAGAUCUACU AAAACAAGGC UUUAUGCCGA AAUCA |
-Macromolecule #3: Target DNA strand
| Macromolecule | Name: Target DNA strand / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.641558 KDa |
| Sequence | String: (DC)(DG)(DC)(DC)(DA)(DC)(DG)(DT)(DC)(DG) (DC)(DC)(DG)(DG)(DA)(DG)(DG)(DA)(DG)(DA) (DG)(DC)(DG)(DA)(DT)(DT)(DA)(DC) |
-Macromolecule #4: Non-target DNA strand
| Macromolecule | Name: Non-target DNA strand / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.467629 KDa |
| Sequence | String: (DA)(DC)(DG)(DT)(DG)(DG)(DC)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Homemade / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 16.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)