+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | LpqY-SugABC in state 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Trehalose / ABC transporter / tuberculosis / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbohydrate transport / maltose binding / maltose transport / maltodextrin transmembrane transport / ABC-type transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / transmembrane transport / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.29 Å | |||||||||
 Authors Authors | Liang J / Yang X / Zhang B / Rao Z / Liu F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC. Authors: Jingxi Liang / Xiuna Yang / Tianyu Hu / Yan Gao / Qi Yang / Haitao Yang / Wei Peng / Xiaoting Zhou / Luke W Guddat / Bing Zhang / Zihe Rao / Fengjiang Liu /   Abstract: The human pathogen, Mycobacterium tuberculosis (Mtb) relies heavily on trehalose for both survival and pathogenicity. The type I ATP-binding cassette (ABC) transporter LpqY-SugABC is the only ...The human pathogen, Mycobacterium tuberculosis (Mtb) relies heavily on trehalose for both survival and pathogenicity. The type I ATP-binding cassette (ABC) transporter LpqY-SugABC is the only trehalose import pathway in Mtb. Conformational dynamics of ABC transporters is an important feature to explain how they operate, but experimental structures are determined in a static environment. Therefore, a detailed transport mechanism cannot be elucidated because there is a lack of intermediate structures. Here, we used single-particle cryo-electron microscopy (cryo-EM) to determine the structure of the Mycobacterium smegmatis (M. smegmatis) trehalose-specific importer LpqY-SugABC complex in five different conformations. These structures have been classified and reconstructed from a single cryo-EM dataset. This study allows a comprehensive understanding of the trehalose recycling mechanism in Mycobacteria and also demonstrates the potential of single-particle cryo-EM to explore the dynamic structures of other ABC transporters and molecular machines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34932.map.gz emd_34932.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34932-v30.xml emd-34932-v30.xml emd-34932.xml emd-34932.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34932.png emd_34932.png | 89 KB | ||
| Masks |  emd_34932_msk_1.map emd_34932_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34932.cif.gz emd-34932.cif.gz | 7.1 KB | ||
| Others |  emd_34932_half_map_1.map.gz emd_34932_half_map_1.map.gz emd_34932_half_map_2.map.gz emd_34932_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34932 http://ftp.pdbj.org/pub/emdb/structures/EMD-34932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34932 | HTTPS FTP |
-Related structure data
| Related structure data |  8hplMC  8hpmC  8hpnC  8hprC  8hpsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34932.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34932.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
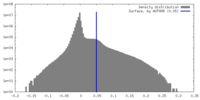
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34932_msk_1.map emd_34932_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
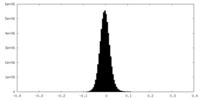
| Density Histograms |
-Half map: #2
| File | emd_34932_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34932_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trehalose-specific importer LpqY-SugABC complex
| Entire | Name: Trehalose-specific importer LpqY-SugABC complex |
|---|---|
| Components |
|
-Supramolecule #1: Trehalose-specific importer LpqY-SugABC complex
| Supramolecule | Name: Trehalose-specific importer LpqY-SugABC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3, #1-#2, #4 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: ABC sugar transporter, permease component
| Macromolecule | Name: ABC sugar transporter, permease component / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 32.73925 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MTAAVTPSAS AVASDDKKSE RRLAFWLIAP AVLLMLAVTA YPIGYAVWLS LQRYNLAEPH DTEFIGLANY VTVLTDGYWW TAFAVTLGI TVVSVAIEFA LGLALALVMH RTIFGKGAVR TAILIPYGIV TVAASYSWYY AWTPGTGYLA NLLPEGSAPL T DQLPSLAI ...String: MTAAVTPSAS AVASDDKKSE RRLAFWLIAP AVLLMLAVTA YPIGYAVWLS LQRYNLAEPH DTEFIGLANY VTVLTDGYWW TAFAVTLGI TVVSVAIEFA LGLALALVMH RTIFGKGAVR TAILIPYGIV TVAASYSWYY AWTPGTGYLA NLLPEGSAPL T DQLPSLAI VVLAEVWKTT PFMALLLLAG LALVPQDLLN AAQVDGAGPW KRLTKVILPM IKPAILVALL FRTLDAFRIF DN IYILTGG SNDTGSVSIL GYDNLFKAFN VGLGSAISVL IFLSVAIIAF IYIKIFGAAA PGSDEEVR UniProtKB: ABC sugar transporter, permease component |
-Macromolecule #2: ABC transporter, permease protein SugB
| Macromolecule | Name: ABC transporter, permease protein SugB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 29.839441 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MADRVDARRA TWWSVVNILV IVYALIPVLW ILSLSLKPTS SVKDGKLIPT EITFANYKAI FSGDAFTSAL FNSIGIGLIT TIIAVVIGG MAAYAVARLQ FPGKQLLIGV ALLIAMFPHI SLVTPIFNMW RGIGLFDTWP GLIIPYITFA LPLAIYTLSA F FREIPWDL ...String: MADRVDARRA TWWSVVNILV IVYALIPVLW ILSLSLKPTS SVKDGKLIPT EITFANYKAI FSGDAFTSAL FNSIGIGLIT TIIAVVIGG MAAYAVARLQ FPGKQLLIGV ALLIAMFPHI SLVTPIFNMW RGIGLFDTWP GLIIPYITFA LPLAIYTLSA F FREIPWDL EKAAKMDGAT PAQAFRKVIA PLAAPGIVTA AILVFIFAWN DLLLALSLTA TQRAITAPVA IANFTGSSQF EE PTGSIAA GAMVITIPII IFVLIFQRRI VAGLTSGAVK G UniProtKB: ABC transporter, permease protein SugB |
-Macromolecule #3: ABC transporter, ATP-binding protein SugC
| Macromolecule | Name: ABC transporter, ATP-binding protein SugC / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 43.702625 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAEIVLDRVT KSYPDGAGGV RAAVKEFSMT IADGEFIILV GPSGCGKSTT LNMIAGLEEI TSGELRIGGE RVNEKAPKDR DIAMVFQSY ALYPHMTVRQ NIAFPLTLAK VPKAEIAAKV EETAKILDLS ELLDRKPGQL SGGQRQRVAM GRAIVRSPKA F LMDQPLSN ...String: MAEIVLDRVT KSYPDGAGGV RAAVKEFSMT IADGEFIILV GPSGCGKSTT LNMIAGLEEI TSGELRIGGE RVNEKAPKDR DIAMVFQSY ALYPHMTVRQ NIAFPLTLAK VPKAEIAAKV EETAKILDLS ELLDRKPGQL SGGQRQRVAM GRAIVRSPKA F LMDQPLSN LDAKLRVQMR AEISRLQDRL GTTTVYVTHD QTEAMTLGDR VVVMLAGEVQ QIGTPDELYS SPANLFVAGF IG SPAMNFF PATRTDVGVR LPFGEVTLTP HMLDLLDKQA RPENIIVGIR PEHIEDSALL DGYARIRALT FSVRADIVES LGA DKYVHF TTEGAGAESA QLAELAADSG AGTNQFIARV SADSRVRTGE QIELAIDTTK LSIFDAATGL NLTRDITPTD PTEA AGPDA G UniProtKB: Trehalose import ATP-binding protein SugC |
-Macromolecule #4: Bacterial extracellular solute-binding protein
| Macromolecule | Name: Bacterial extracellular solute-binding protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 50.183086 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MRARRLCAAA VAAMAAASMV SACGSQTGGI VINYYTPANE EATFKAVANR CNEQLGGRFQ IAQRNLPKGA DDQRLQLARR LTGNDKSLD VMALDVVWTA EFAEAGWAVP LSEDPAGLAE ADATENTLPG PLETARWQDE LYAAPITTNT QLLWYRADLM P APPTTWDG ...String: MRARRLCAAA VAAMAAASMV SACGSQTGGI VINYYTPANE EATFKAVANR CNEQLGGRFQ IAQRNLPKGA DDQRLQLARR LTGNDKSLD VMALDVVWTA EFAEAGWAVP LSEDPAGLAE ADATENTLPG PLETARWQDE LYAAPITTNT QLLWYRADLM P APPTTWDG MLDEANRLYR EGGPSWIAVQ GKQYEGMVVW FNTLLQSAGG QVLSDDGQRV TLTDTPEHRA ATVKALRIIK SV ATAPGAD PSITQTDENT ARLALEQGKA ALEVNWPYVL PSLLENAVKG GVSFLPLDGD PALQGSINDV GTFSPTDEQF DIA FDASKK VFGFAPYPGV NPDEPARVTL GGLNLAVAST SQHKAEAFEA IRCLRNVENQ RYTSIEGGLP AVRTSLYDDP AFQK KYPQY EIIRQQLTNA AVRPATPVYQ AVSTRMSATL APISDIDPER TADELTEAVQ KAIDGKGLIP UniProtKB: Bacterial extracellular solute-binding protein |
-Macromolecule #6: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)