[English] 日本語
 Yorodumi
Yorodumi- EMDB-34904: Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pre-tRNA splicing / TSEN / pre-tRNA / RNA BINDING PROTEIN/RNA / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA-intron endonuclease complex / tRNA-type intron splice site recognition and cleavage / ATP-dependent polyribonucleotide 5'-hydroxyl-kinase activity / polynucleotide 5'-hydroxyl-kinase / tRNA-intron lyase / tRNA-intron lyase activity / ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity / polynucleotide 5'-hydroxyl-kinase activity / mRNA cleavage factor complex / global gene silencing by mRNA cleavage ...tRNA-intron endonuclease complex / tRNA-type intron splice site recognition and cleavage / ATP-dependent polyribonucleotide 5'-hydroxyl-kinase activity / polynucleotide 5'-hydroxyl-kinase / tRNA-intron lyase / tRNA-intron lyase activity / ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity / polynucleotide 5'-hydroxyl-kinase activity / mRNA cleavage factor complex / global gene silencing by mRNA cleavage / tRNA splicing, via endonucleolytic cleavage and ligation / cerebellar cortex development / Processing of Intronless Pre-mRNAs / RISC complex assembly / mRNA 3'-end processing / mRNA 3'-end processing / tRNA processing in the nucleus / RNA Polymerase II Transcription Termination / tRNA processing / Processing of Capped Intron-Containing Pre-mRNA / mRNA processing / nucleic acid binding / lyase activity / centrosome / nucleolus / nucleoplasm / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Zhang X / Yang F / Zhan X / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease. Authors: Xiaofeng Zhang / Fenghua Yang / Xiechao Zhan / Tong Bian / Zhihan Xing / Yichen Lu / Yigong Shi /  Abstract: Removal of the intron from precursor-tRNA (pre-tRNA) is essential in all three kingdoms of life. In humans, this process is mediated by the tRNA splicing endonuclease (TSEN) comprising four subunits: ...Removal of the intron from precursor-tRNA (pre-tRNA) is essential in all three kingdoms of life. In humans, this process is mediated by the tRNA splicing endonuclease (TSEN) comprising four subunits: TSEN2, TSEN15, TSEN34, and TSEN54. Here, we report the cryo-EM structures of human TSEN bound to full-length pre-tRNA in the pre-catalytic and post-catalytic states at average resolutions of 2.94 and 2.88 Å, respectively. Human TSEN features an extended surface groove that holds the L-shaped pre-tRNA. The mature domain of pre-tRNA is recognized by conserved structural elements of TSEN34, TSEN54, and TSEN2. Such recognition orients the anticodon stem of pre-tRNA and places the 3'-splice site and 5'-splice site into the catalytic centers of TSEN34 and TSEN2, respectively. The bulk of the intron sequences makes no direct interaction with TSEN, explaining why pre-tRNAs of varying introns can be accommodated and cleaved. Our structures reveal the molecular ruler mechanism of pre-tRNA cleavage by TSEN. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34904.map.gz emd_34904.map.gz | 49.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34904-v30.xml emd-34904-v30.xml emd-34904.xml emd-34904.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34904.png emd_34904.png | 62.1 KB | ||
| Filedesc metadata |  emd-34904.cif.gz emd-34904.cif.gz | 7.7 KB | ||
| Others |  emd_34904_half_map_1.map.gz emd_34904_half_map_1.map.gz emd_34904_half_map_2.map.gz emd_34904_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34904 http://ftp.pdbj.org/pub/emdb/structures/EMD-34904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34904 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmyMC  8hmzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34904.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34904.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
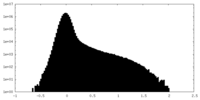
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34904_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
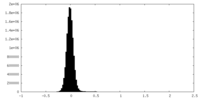
| Density Histograms |
-Half map: #1
| File | emd_34904_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
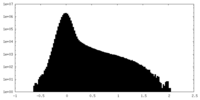
| Density Histograms |
- Sample components
Sample components
-Entire : The human pre-catalytic TSEN/pre-tRNA complex
| Entire | Name: The human pre-catalytic TSEN/pre-tRNA complex |
|---|---|
| Components |
|
-Supramolecule #1: The human pre-catalytic TSEN/pre-tRNA complex
| Supramolecule | Name: The human pre-catalytic TSEN/pre-tRNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: tRNA-splicing endonuclease subunit Sen2
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNA-intron lyase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.391945 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASDYKDDDD KASDEVDAGT MAEAVFHAPK RKRRVYETYE SPLPIPFGQD HGPLKEFKIF RAEMINNNVI VRNAEDIEQL YGKGYFGKG ILSRSRPSFT ISDPKLVAKW KDMKTNMPII TSKRYQHSVE WAAELMRRQG QDESTVRRIL KDYTKPLEHP P VKRNEEAQ ...String: MASDYKDDDD KASDEVDAGT MAEAVFHAPK RKRRVYETYE SPLPIPFGQD HGPLKEFKIF RAEMINNNVI VRNAEDIEQL YGKGYFGKG ILSRSRPSFT ISDPKLVAKW KDMKTNMPII TSKRYQHSVE WAAELMRRQG QDESTVRRIL KDYTKPLEHP P VKRNEEAQ VHDKLNSGMV SNMEGTAGGE RPSVVNGDSG KSGGVGDPRE PLGCLQEGSG CHPTTESFEK SVREDASPLP HV CCCKQDA LILQRGLHHE DGSQHIGLLH PGDRGPDHEY VLVEEAECAM SEREAAPNEE LVQRNRLICR RNPYRIFEYL QLS LEEAFF LVYALGCLSI YYEKEPLTIV KLWKAFTVVQ PTFRTTYMAY HYFRSKGWVP KVGLKYGTDL LLYRKGPPFY AASY SVIIE LVDDHFEGSL RRPLSWKSLA ALSRVSVNVS KELMLCYLIK PSTMTDKEME SPECMKRIKV QEVILSRWVS SRERS DQDD L UniProtKB: tRNA-splicing endonuclease subunit Sen2 |
-Macromolecule #2: tRNA-splicing endonuclease subunit Sen34
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen34 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNA-intron lyase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.759938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASDYKDDDD KASDEVDAGT MLVVEVANGR SLVWGAEAVQ ALRERLGVGG RTVGALPRGP RQNSRLGLPL LLMPEEARLL AEIGAVTLV SAPRPDSRHH SLALTSFKRQ QEESFQEQSA LAAEARETRR QELLEKITEG QAAKKQKLEQ ASGASSSQEA G SSQAAKED ...String: MASDYKDDDD KASDEVDAGT MLVVEVANGR SLVWGAEAVQ ALRERLGVGG RTVGALPRGP RQNSRLGLPL LLMPEEARLL AEIGAVTLV SAPRPDSRHH SLALTSFKRQ QEESFQEQSA LAAEARETRR QELLEKITEG QAAKKQKLEQ ASGASSSQEA G SSQAAKED ETSDGQASGE QEEAGPSSSQ AGPSNGVAPL PRSALLVQLA TARPRPVKAR PLDWRVQSKD WPHAGRPAHE LR YSIYRDL WERGFFLSAA GKFGGDFLVY PGDPLRFAAH YIAQCWAPED TIPLQDLVAA GRLGTSVRKT LLLCSPQPDG KVV YTSLQW ASLQ UniProtKB: tRNA-splicing endonuclease subunit Sen34 |
-Macromolecule #4: tRNA-splicing endonuclease subunit Sen54
| Macromolecule | Name: tRNA-splicing endonuclease subunit Sen54 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.03234 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASDYKDDDD KASDEVDAGT MEPEPEPAAV EVPAGRVLSA RELFAARSRS QKLPQRSHGP KDFLPDGSAA QAERLRRCRE ELWQLLAEQ RVERLGSLVA AEWRPEEGFV ELKSPAGKFW QTMGFSEQGR QRLHPEEALY LLECGSIHLF HQDLPLSIQE A YQLLLTDH ...String: MASDYKDDDD KASDEVDAGT MEPEPEPAAV EVPAGRVLSA RELFAARSRS QKLPQRSHGP KDFLPDGSAA QAERLRRCRE ELWQLLAEQ RVERLGSLVA AEWRPEEGFV ELKSPAGKFW QTMGFSEQGR QRLHPEEALY LLECGSIHLF HQDLPLSIQE A YQLLLTDH TVTFLQYQVF SHLKRLGYVV RRFQPSSVLS PYERQLNLDA SVQHLEDGDG KRKRSSSSPR SINKKAKALD NS LQPKSLA ASSPPPCSQP SQCPEEKPQE SSPMKGPGGP FQLLGSLGPS PGPAREGVGC SWESGRAENG VTGAGKRRWN FEQ ISFPNM ASDSRHTLLR APAPELLPAN VAGRETDAES WCQKLNQRKE KLSRREREHH AEAAQFQEDV NADPEVQRCS SWRE YKELL QRRQVQRSQR RAPHLWGQPV TPLLSPGQAS SPAVVLQHIS VLQTTHLPDG GARLLEKSGG LEIIFDVYQA DAVAT FRKN NPGKPYARMC ISGFDEPVPD LCSLKRLSYQ SGDVPLIFAL VDHGDISFYS FRDFTLPQDV GH UniProtKB: tRNA-splicing endonuclease subunit Sen54 |
-Macromolecule #5: Chromosome 1 open reading frame 19, isoform CRA_a
| Macromolecule | Name: Chromosome 1 open reading frame 19, isoform CRA_a / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.796555 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASSAWSHPQ FEKGGGSGGG SGGSAWSHPQ FEKGSAAAME ERGDSEPTPG CSGLGPGGVR GFGDGGGAPS WAPEDAWMGT HPKYLEMME LDIGDATQVY VAFLVYLDLM ESKSWHEVNC VGLPELQLIC LVGTEIEGEG LQTVVPTPIT ASLSHNRIRE I LKASRKLQ ...String: MASSAWSHPQ FEKGGGSGGG SGGSAWSHPQ FEKGSAAAME ERGDSEPTPG CSGLGPGGVR GFGDGGGAPS WAPEDAWMGT HPKYLEMME LDIGDATQVY VAFLVYLDLM ESKSWHEVNC VGLPELQLIC LVGTEIEGEG LQTVVPTPIT ASLSHNRIRE I LKASRKLQ GDPDLPMSFT LAIVESDSTI VYYKLTDGFM LPDPQVSFEN ISLRR UniProtKB: tRNA-splicing endonuclease subunit SEN15 |
-Macromolecule #6: Polyribonucleotide 5'-hydroxyl-kinase Clp1
| Macromolecule | Name: Polyribonucleotide 5'-hydroxyl-kinase Clp1 / type: protein_or_peptide / ID: 6 Details: Author stated that the EM density map for the chain E is relatively weak and could be assigned in the low resolution map. Number of copies: 1 / Enantiomer: LEVO / EC number: polynucleotide 5'-hydroxyl-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.837504 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASDYKDDDD KASDEVDAGT MGEEANDDKK PTTKFELERE TELRFEVEAS QSVQLELLTG MAEIFGTELT RNKKFTFDAG AKVAVFTWH GCSVQLSGRT EVAYVSKDTP MLLYLNTHTA LEQMRRQAEK EEERGPRVMV VGPTDVGKST VCRLLLNYAV R LGRRPTYV ...String: MASDYKDDDD KASDEVDAGT MGEEANDDKK PTTKFELERE TELRFEVEAS QSVQLELLTG MAEIFGTELT RNKKFTFDAG AKVAVFTWH GCSVQLSGRT EVAYVSKDTP MLLYLNTHTA LEQMRRQAEK EEERGPRVMV VGPTDVGKST VCRLLLNYAV R LGRRPTYV ELDVGQGSVS IPGTMGALYI ERPADVEEGF SIQAPLVYHF GSTTPGTNIK LYNKITSRLA DVFNQRCEVN RR ASVSGCV INTCGWVKGS GYQALVHAAS AFEVDVVVVL DQERLYNELK RDLPHFVRTV LLPKSGGVVE RSKDFRRECR DER IREYFY GFRGCFYPHA FNVKFSDVKI YKVGAPTIPD SCLPLGMSQE DNQLKLVPVT PGRDMVHHLL SVSTAEGTEE NLSE TSVAG FIVVTSVDLE HQVFTVLSPA PRPLPKNFLL IMDIRFMDLK UniProtKB: Polyribonucleotide 5'-hydroxyl-kinase Clp1 |
-Macromolecule #3: Pre-tRNA
| Macromolecule | Name: Pre-tRNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.798828 KDa |
| Sequence | String: UAAUACGACU CACUAUAGGG GGCUCUGUGG CGCAAUGGAU AGCGCAUUGG ACUUCUAGUG ACGAAUAGAG CAAUUCAAAG GUUGUGGGU UCGAAUCCCA CCAGAGUCGG AUAUC |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)