+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GPR52 with Gs and c17 | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled photoreceptor activity / cellular response to light stimulus / PKA activation in glucagon signalling / phototransduction / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis ...G protein-coupled photoreceptor activity / cellular response to light stimulus / PKA activation in glucagon signalling / phototransduction / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / locomotory behavior / bone development / G protein-coupled receptor activity / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / response to xenobiotic stimulus / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.77 Å | |||||||||
 Authors Authors | Chen B / Xu F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: MedComm (2020) / Year: 2023 Journal: MedComm (2020) / Year: 2023Title: Allosteric coupling between G-protein binding and extracellular ligand binding sites in GPR52 revealed by F-NMR and cryo-electron microscopy. Authors: Yanliu Fan / Xi Lin / Benxun Pan / Bo Chen / Dongsheng Liu / Kurt Wüthrich / Fei Xu /    | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34902.map.gz emd_34902.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34902-v30.xml emd-34902-v30.xml emd-34902.xml emd-34902.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34902_fsc.xml emd_34902_fsc.xml | 17.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34902.png emd_34902.png | 43.6 KB | ||
| Filedesc metadata |  emd-34902.cif.gz emd-34902.cif.gz | 6.9 KB | ||
| Others |  emd_34902_half_map_1.map.gz emd_34902_half_map_1.map.gz emd_34902_half_map_2.map.gz emd_34902_half_map_2.map.gz | 387.1 MB 387.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34902 http://ftp.pdbj.org/pub/emdb/structures/EMD-34902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34902 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34902.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34902.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.55467 Å | ||||||||||||||||||||||||||||||||||||
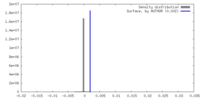
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1 map
| File | emd_34902_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
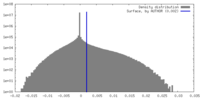
| Density Histograms |
-Half map: half2 map
| File | emd_34902_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPR52 and Gs with C17
| Entire | Name: GPR52 and Gs with C17 |
|---|---|
| Components |
|
-Supramolecule #1: GPR52 and Gs with C17
| Supramolecule | Name: GPR52 and Gs with C17 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.366764 KDa |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: HHHHHHGGGS DSLEFIASKL AGGGSMASNN TASIAQARKL VEQLKMEANI DRIKVSKAAA DLMAYCEAHA KEDPLLTPVP ASENPFREK KFFSAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #3: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.054232 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMQVQLQES GGGLVQPGGS LRLSCAASGF TFSNYKMNWV RQAPGKGLEW VSDISQSGAS ISYTGSVKG RFTISRDNAK NTLYLQMNSL KPEDTAVYYC ARCPAPFTRD CFDVTSTTYA YRGQGTQVTV |
-Macromolecule #4: G-protein coupled receptor 52
| Macromolecule | Name: G-protein coupled receptor 52 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.28652 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MNESRWTEWR ILNMSSGIVN VSERHSCPLG FGHYSVVDVC IFETVVIVLL TFLIIAGNLT VIFVFHCAPL LHHYTTSYFI QTMAYADLF VGVSCLVPTL SLLHYSTGVH ESLTCQVFGY IISVLKSVSM WCLACISVDR YLAITKPLSY NQLVTPCRLR I CIILIWIY ...String: MNESRWTEWR ILNMSSGIVN VSERHSCPLG FGHYSVVDVC IFETVVIVLL TFLIIAGNLT VIFVFHCAPL LHHYTTSYFI QTMAYADLF VGVSCLVPTL SLLHYSTGVH ESLTCQVFGY IISVLKSVSM WCLACISVDR YLAITKPLSY NQLVTPCRLR I CIILIWIY SCLIFLPSFF GWGKPGYHGD IFEWCATSWL TSAYFTGFIV CLLYAPAAFV VCFTYFHIFK ICRQHTKEIN DR RARFPSH EVDSSRETGH SPDRRYAMVL FRITSVFYML WLPYIIYFLL ESSRVLDNPT LSFLTTWLAI SNSFCNPVIY SLS NSVF UniProtKB: G-protein coupled receptor 52 |
-Macromolecule #5: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.964734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKIEDYFP ...String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKIEDYFP EFARYTTPED ATPEPGEDPR VTRAKYFIRD EFLRISTASG DGRHYCYPHF TCAVDTENAR RIFNDCRDII QR MHLRQYE LL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #6: N-(2-hydroxyethyl)-5-(hydroxymethyl)-3-methyl-1-[2-[[3-(trifluoro...
| Macromolecule | Name: N-(2-hydroxyethyl)-5-(hydroxymethyl)-3-methyl-1-[2-[[3-(trifluoromethyl)phenyl]methyl]-1-benzothiophen-7-yl]pyrazole-4-carboxamide type: ligand / ID: 6 / Number of copies: 1 / Formula: EN6 |
|---|---|
| Molecular weight | Theoretical: 489.51 Da |
| Chemical component information |  ChemComp-EN6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)