[English] 日本語
 Yorodumi
Yorodumi- EMDB-34892: Cryo-EM structure of human high-voltage activated L-type calcium ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ben bound state / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels / positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during atrial cardiac muscle cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase / photoreceptor ribbon synapse / membrane depolarization during AV node cell action potential / membrane depolarization during bundle of His cell action potential ...voltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels / positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during atrial cardiac muscle cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase / photoreceptor ribbon synapse / membrane depolarization during AV node cell action potential / membrane depolarization during bundle of His cell action potential / signaling / L-type voltage-gated calcium channel complex / cell communication / positive regulation of muscle contraction / NCAM1 interactions / regulation of ventricular cardiac muscle cell membrane repolarization / positive regulation of calcium ion transport / cardiac muscle cell action potential involved in contraction / calcium ion import / calcium ion transport into cytosol / Sensory processing of sound by inner hair cells of the cochlea / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / voltage-gated calcium channel complex / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / calcium ion transmembrane import into cytosol / neuromuscular junction development / regulation of heart contraction / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / calcium ion import across plasma membrane / regulation of calcium ion transport / neuronal dense core vesicle / voltage-gated calcium channel activity / presynaptic active zone membrane / visual perception / T-tubule / muscle contraction / sarcoplasmic reticulum / protein localization to plasma membrane / regulation of membrane potential / Regulation of insulin secretion / postsynaptic density membrane / calcium ion transmembrane transport / GABA-ergic synapse / calcium channel activity / cellular response to amyloid-beta / actin filament binding / Adrenaline,noradrenaline inhibits insulin secretion / calcium ion transport / presynapse / perikaryon / chemical synaptic transmission / calmodulin binding / cilium / dendrite / extracellular exosome / nucleoplasm / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
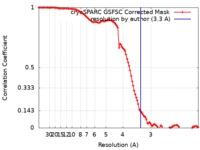
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Wei Y / Yu Z / Zhao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural bases of inhibitory mechanism of Ca1.2 channel inhibitors. Authors: Yiqing Wei / Zhuoya Yu / Lili Wang / Xiaojing Li / Na Li / Qinru Bai / Yuhang Wang / Renjie Li / Yufei Meng / Hao Xu / Xianping Wang / Yanli Dong / Zhuo Huang / Xuejun Cai Zhang / Yan Zhao /  Abstract: The voltage-gated calcium channel Ca1.2 is essential for cardiac and vessel smooth muscle contractility and brain function. Accumulating evidence demonstrates that malfunctions of Ca1.2 are involved ...The voltage-gated calcium channel Ca1.2 is essential for cardiac and vessel smooth muscle contractility and brain function. Accumulating evidence demonstrates that malfunctions of Ca1.2 are involved in brain and heart diseases. Pharmacological inhibition of Ca1.2 is therefore of therapeutic value. Here, we report cryo-EM structures of Ca1.2 in the absence or presence of the antirheumatic drug tetrandrine or antihypertensive drug benidipine. Tetrandrine acts as a pore blocker in a pocket composed of S6, S6, and S6 helices and forms extensive hydrophobic interactions with Ca1.2. Our structure elucidates that benidipine is located in the D-D fenestration site. Its hydrophobic sidechain, phenylpiperidine, is positioned at the exterior of the pore domain and cradled within a hydrophobic pocket formed by S5, S6, and S6 helices, providing additional interactions to exert inhibitory effects on both L-type and T-type voltage gated calcium channels. These findings provide the structural foundation for the rational design and optimization of therapeutic inhibitors of voltage-gated calcium channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34892.map.gz emd_34892.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34892-v30.xml emd-34892-v30.xml emd-34892.xml emd-34892.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34892_fsc.xml emd_34892_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34892.png emd_34892.png | 41.4 KB | ||
| Filedesc metadata |  emd-34892.cif.gz emd-34892.cif.gz | 9 KB | ||
| Others |  emd_34892_half_map_1.map.gz emd_34892_half_map_1.map.gz emd_34892_half_map_2.map.gz emd_34892_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34892 http://ftp.pdbj.org/pub/emdb/structures/EMD-34892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34892 | HTTPS FTP |
-Related structure data
| Related structure data |  8hmbMC  8hlpC  8hmaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34892.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34892.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34892_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
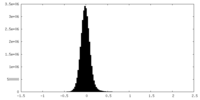
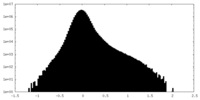
| Density Histograms |
-Half map: #1
| File | emd_34892_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
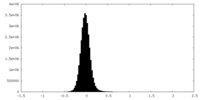
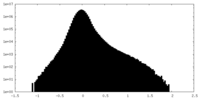
| Density Histograms |
- Sample components
Sample components
-Entire : CaV3.3
| Entire | Name: CaV3.3 |
|---|---|
| Components |
|
-Supramolecule #1: CaV3.3
| Supramolecule | Name: CaV3.3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform 2c of Voltage-dependent L-type calcium channel subunit beta-2
| Macromolecule | Name: Isoform 2c of Voltage-dependent L-type calcium channel subunit beta-2 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.952688 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNQGSGLDLL KISYGKGARR KNRFKGSDGS TSSDTTSNSF VRQGSADSYT SRPSDSDVSL EEDREAVRRE AERQAQAQLE KAKTKPVAF AVRTNVSYSA AHEDDVPVPG MAISFEAKDF LHVKEKFNND WWIGRLVKEG CEIGFIPSPV KLENMRLQHE Q RAKQGKFY ...String: MNQGSGLDLL KISYGKGARR KNRFKGSDGS TSSDTTSNSF VRQGSADSYT SRPSDSDVSL EEDREAVRRE AERQAQAQLE KAKTKPVAF AVRTNVSYSA AHEDDVPVPG MAISFEAKDF LHVKEKFNND WWIGRLVKEG CEIGFIPSPV KLENMRLQHE Q RAKQGKFY SSKSGGNSSS SLGDIVPSSR KSTPPSSAID IDATGLDAEE NDIPANHRSP KPSANSVTSP HSKEKRMPFF KK TEHTPPY DVVPSMRPVV LVGPSLKGYE VTDMMQKALF DFLKHRFEGR ISITRVTADI SLAKRSVLNN PSKHAIIERS NTR SSLAEV QSEIERIFEL ARTLQLVVLD ADTINHPAQL SKTSLAPIIV YVKISSPKVL QRLIKSRGKS QAKHLNVQMV AADK LAQCP PELFDVILDE NQLEDACEHL ADYLEAYWKA THPPSSSLPN PLLSRTLATS SLPLSPTLAS NSQGSQGDQR TDRSA PIRS ASQAEEEPSV EPVKKSQHRS SSSAPHHNHR SGTSRGLSRQ ETFDSETQES RDSAYVEPKE DYSHDHVDHY ASHRDH NHR DETHGSSDHR HRESRHRSRD VDREQDHNEC NKQRSRHKSK DRYCEKDGEV ISKKRNEAGE WNRDVYIRQ UniProtKB: Voltage-dependent L-type calcium channel subunit beta-2 |
-Macromolecule #2: Voltage-dependent L-type calcium channel subunit alpha
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit alpha type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 243.947016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVNENTRMYI PEENHQGSNY GSPRPAHANM NANAAAGLAP EHIPTPGAAL SWQAAIDAAR QAKLMGSAGN ATISTVSSTQ RKRQQYGKP KKQGSTTATR PPRALLCLTL KNPIRRACIS IVEWKPFEII ILLTIFANCV ALAIYIPFPE DDSNATNSNL E RVEYLFLI ...String: MVNENTRMYI PEENHQGSNY GSPRPAHANM NANAAAGLAP EHIPTPGAAL SWQAAIDAAR QAKLMGSAGN ATISTVSSTQ RKRQQYGKP KKQGSTTATR PPRALLCLTL KNPIRRACIS IVEWKPFEII ILLTIFANCV ALAIYIPFPE DDSNATNSNL E RVEYLFLI IFTVEAFLKV IAYGLLFHPN AYLRNGWNLL DFIIVVVGLF SAILEQATKA DGANALGGKG AGFDVKALRA FR VLRPLRL VSGVPSLQVV LNSIIKAMVP LLHIALLVLF VIIIYAIIGL ELFMGKMHKT CYNQEGIADV PAEDDPSPCA LET GHGRQC QNGTVCKPGW DGPKHGITNF DNFAFAMLTV FQCITMEGWT DVLYWVNDAV GRDWPWIYFV TLIIIGSFFV LNLV LGVLS GEFSKEREKA KARGDFQKLR EKQQLEEDLK GYLDWITQAE DIDPENEDEG MDEEKPRNMS MPTSETESVN TENVA GGDI EGENCGARLA HRISKSKFSR YWRRWNRFCR RKCRAAVKSN VFYWLVIFLV FLNTLTIASE HYNQPNWLTE VQDTAN KAL LALFTAEMLL KMYSLGLQAY FVSLFNRFDC FVVCGGILET ILVETKIMSP LGISVLRCVR LLRIFKITRY WNSLSNL VA SLLNSVRSIA SLLLLLFLFI IIFSLLGMQL FGGKFNFDEM QTRRSTFDNF PQSLLTVFQI LTGEDWNSVM YDGIMAYG G PSFPGMLVCI YFIILFICGN YILLNVFLAI AVDNLADAES LTSAQKEEEE EKERKKLART ASPEKKQELV EKPAVGESK EEKIELKSIT ADGESPPATK INMDDLQPNE NEDKSPYPNP ETTGEEDEEE PEMPVGPRPR PLSELHLKEK AVPMPEASAF FIFSSNNRF RLQCHRIVND TIFTNLILFF ILLSSISLAA EDPVQHTSFR NHILFYFDIV FTTIFTLEII LKMTAYGAFL H KGSFCRNY FNILDLLVVS VSLISFGIQS SAINVVKILR VLRVLRPLRA INRAKGLKHV VQCVFVAIRT IGNIVIVTTL LQ FMFACIG VQLFKGKLYT CSDSSKQTEA ECKGNYITYK DGEVDHPIIQ PRSWENSKFD FDNVLAAMMA LFTVSTFEGW PEL LYRSID SHTEDKGPIY NYRVEISIFF IIYIIIIAFF MMNIFVGFVI VTFQEQGEQE YKNCELDKNQ RQCVEYALKA RPLR RYIPK NQHQYKVWYV VNSTYFEYLM FVLILLNTIC LAMQHYGQSC LFKIAMNILN MLFTGLFTVE MILKLIAFKP KGYFS DPWN VFDFLIVIGS IIDVILSEVN PAEHTQCSPS MNAEENSRIS ITFFRLFRVM RLVKLLSRGE GIRTLLWTFI KSFQAL PYV ALLIVMLFFI YAVIGMQVFG KIALNDTTEI NRNNNFQTFP QAVLLLFRCA TGEAWQDIML ACMPGKKCAP ESEPSNS TE GETPCGSSFA VFYFISFYML CAFLIINLFV AVIMDNFDYL TRDWSILGPH HLDEFKRIWA EYDPEAKGRI KHLDVVTL L RRIQPPLGFG KLCPHRVACK RLVSMNMPLN SDGTVMFNAT LFALVRTALR IKTEGNLEQA NEELRAIIKK IWKRTSMKL LDQVVPPAGD DEVTVGKFYA TFLIQEYFRK FKKRKEQGLV GKPSQRNALS LQAGLRTLHD IGPEIRRAIS GDLTAEEELD KAMKEAVSA ASEDDIFRRA GGLFGNHVSY YQSDGRSAFP QTFTTQRPLH INKAGSSQGD TESPSHEKLV DSTFTPSSYS S TGSNANIN NANNTALGRL PRPAGYPSTV STVEGHGPPL SPAIRVQEVA WKLSSNRERH VPMCEDLELR RDSGSAGTQA HC LLLRRAN PSRCHSRESQ AAMAGQEETS QDETYEVKMN HDTEACSEPS LLSTEMLSYQ DDENRQLTLP EEDKRDIRQS PKR GFLRSA SLGRRASFHL ECLKRQKDRG GDISQKTVLP LHLVHHQALA VAGLSPLLQR SHSPASFPRP FATPPATPGS RGWP PQPVP TLRLEGVESS EKLNSSFPSI HCGSWAETTP GGGGSSAARR VRPVSLMVPS QAGAPGRQFH GSASSLVEAV LISEG LGQF AQDPKFIEVT TQELADACDM TIEEMESAAD NILSGGAPQS PNGALLPFVN CRDAGQDRAG GEEDAGCVRA RGRPSE EEL QDSRVYVSSL UniProtKB: Voltage-dependent L-type calcium channel subunit alpha |
-Macromolecule #3: Voltage-dependent calcium channel subunit alpha-2/delta-1
| Macromolecule | Name: Voltage-dependent calcium channel subunit alpha-2/delta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 121.197406 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI ...String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI PTDIYEGSTI VLNELNWTSA LDEVFKKNRE EDPSLLWQVF GSATGLARYY PASPWVDNSR TPNKIDLYDV RR RPWYIQG AASPKDMLIL VDVSGSVSGL TLKLIRTSVS EMLETLSDDD FVNVASFNSN AQDVSCFQHL VQANVRNKKV LKD AVNNIT AKGITDYKKG FSFAFEQLLN YNVSRANCNK IIMLFTDGGE ERAQEIFNKY NKDKKVRVFT FSVGQHNYDR GPIQ WMACE NKGYYYEIPS IGAIRINTQE YLDVLGRPMV LAGDKAKQVQ WTNVYLDALE LGLVITGTLP VFNITGQFEN KTNLK NQLI LGVMGVDVSL EDIKRLTPRF TLCPNGYYFA IDPNGYVLLH PNLQPKPIGV GIPTINLRKR RPNIQNPKSQ EPVTLD FLD AELENDIKVE IRNKMIDGES GEKTFRTLVK SQDERYIDKG NRTYTWTPVN GTDYSLALVL PTYSFYYIKA KLEETIT QA RYSETLKPDN FEESGYTFIA PRDYCNDLKI SDNNTEFLLN FNEFIDRKTP NNPSCNADLI NRVLLDAGFT NELVQNYW S KQKNIKGVKA RFVVTDGGIT RVYPKEAGEN WQENPETYED SFYKRSLDND NYVFTAPYFN KSGPGAYESG IMVSKAVEI YIQGKLLKPA VVGIKIDVNS WIENFTKTSI RDPCAGPVCD CKRNSDVMDC VILDDGGFLL MANHDDYTNQ IGRFFGEIDP SLMRHLVNI SVYAFNKSYD YQSVCEPGAA PKQGAGHRSA YVPSVADILQ IGWWATAAAW SILQQFLLSL TFPRLLEAVE M EDDDFTAS LSKQSCITEQ TQYFFDNDSK SFSGVLDCGN CSRIFHGEKL MNTNLIFIMV ESKGTCPCDT RLLIQAEQTS DG PNPCDMV KQPRYRKGPD VCFDNNVLED YTDC UniProtKB: Voltage-dependent calcium channel subunit alpha-2/delta-1 |
-Macromolecule #4: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 4 / Number of copies: 8 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Macromolecule #5: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 5 / Number of copies: 3 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #6: (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-...
| Macromolecule | Name: (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate type: ligand / ID: 6 / Number of copies: 1 / Formula: A1AC8 |
|---|---|
| Molecular weight | Theoretical: 506.57 Da |
-Macromolecule #7: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 9.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)