+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Echovirus3 empty particle in complex with 6D10 Fab (upright) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E3 Full particle / Echovirus3 Full particle / antibody / 6D10 / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / viral capsid / nucleoside-triphosphate phosphatase ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / viral capsid / nucleoside-triphosphate phosphatase / host cell / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   Echovirus E3 Echovirus E3 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Wang X / Fu W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody. Authors: Shuai Qi / Wangjun Fu / Jinyan Fan / Li Zhang / Binyang Zheng / Kang Wang / Xiangxi Wang / Ling Zhu / Xinjian Li / Yuxia Zhang /  Abstract: Echovirus 3 (E3), a serotype of human enterovirus B (HEV-B), causes severe diseases in infants. Here, we determined the structures of E3 with a monoclonal antibody (MAb) 6D10 by cryo-EM to ...Echovirus 3 (E3), a serotype of human enterovirus B (HEV-B), causes severe diseases in infants. Here, we determined the structures of E3 with a monoclonal antibody (MAb) 6D10 by cryo-EM to comprehensively understand the specificities and the immunological characteristic of this serotype. The solved cryo-EM structures of the F-, A-, and E-particles of E3 bound with 6D10 revealed the structural features of the virus-antibody interface. Importantly, the structures of E-particles bound with 6D10 revealed for the first time the nature of the C-terminus of VP1 for HEV-Bs at the structural level. The highly immunogenic nature of this region in the E-particles provides new strategies for vaccine development for HEV-Bs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34233.map.gz emd_34233.map.gz | 196.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34233-v30.xml emd-34233-v30.xml emd-34233.xml emd-34233.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34233.png emd_34233.png | 55.5 KB | ||
| Filedesc metadata |  emd-34233.cif.gz emd-34233.cif.gz | 6.4 KB | ||
| Others |  emd_34233_half_map_1.map.gz emd_34233_half_map_1.map.gz emd_34233_half_map_2.map.gz emd_34233_half_map_2.map.gz | 166.1 MB 165.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34233 http://ftp.pdbj.org/pub/emdb/structures/EMD-34233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34233 | HTTPS FTP |
-Related structure data
| Related structure data |  8gseMC  8gscC  8gsdC  8gsfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34233.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34233.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34233_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
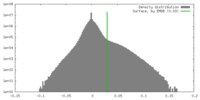
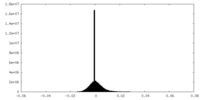
| Density Histograms |
-Half map: #2
| File | emd_34233_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
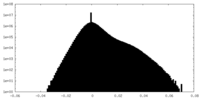
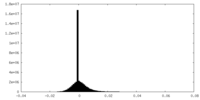
| Density Histograms |
- Sample components
Sample components
-Entire : Echovirus3 empty particle in complex with 6D10 Fab (upright)
| Entire | Name: Echovirus3 empty particle in complex with 6D10 Fab (upright) |
|---|---|
| Components |
|
-Supramolecule #1: Echovirus3 empty particle in complex with 6D10 Fab (upright)
| Supramolecule | Name: Echovirus3 empty particle in complex with 6D10 Fab (upright) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #3: 6D10 Fab
| Supramolecule | Name: 6D10 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Echovirus E3
| Supramolecule | Name: Echovirus E3 / type: virus / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 / NCBI-ID: 47516 / Sci species name: Echovirus E3 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E3 Echovirus E3 |
| Molecular weight | Theoretical: 26.940322 KDa |
| Sequence | String: YHSRTESSIE NFLCRAACVY IATYASAGGT PTERYASWRI NTRQMVQLRR KFELFTYLRF DMEITFVITS TQDPGTQLAQ DMPVLTHQI MYIPPGGPVP NSATDFAWQS STNPSIFWTE GCAPARMSVP FISIGNAYSN FYDGWSHFTQ EGVYGFNSLN N MGHIYVRH ...String: YHSRTESSIE NFLCRAACVY IATYASAGGT PTERYASWRI NTRQMVQLRR KFELFTYLRF DMEITFVITS TQDPGTQLAQ DMPVLTHQI MYIPPGGPVP NSATDFAWQS STNPSIFWTE GCAPARMSVP FISIGNAYSN FYDGWSHFTQ EGVYGFNSLN N MGHIYVRH VNEQSLGVST STLRVYFKPK HVRAWVPRPP RLSPYVKSSN VNFKPTAVTT ERKDINDVGT LRPVGYTNH UniProtKB: Genome polyprotein |
-Macromolecule #2: VP0
| Macromolecule | Name: VP0 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E3 Echovirus E3 |
| Molecular weight | Theoretical: 28.069537 KDa |
| Sequence | String: RVRSITLGNS TITTQECANV VVGYGVWPSY LQDNEATAED QPTQPDVATC RFYTLDSIQW QKESDGWWWK FPEALKNMGL FGQNMEYHY LGRSGYTIHV QCNASKFHQG CLLVVCVPEA EMGCSDVERE VVAASLSSED TAKSFSRTES NGQHTVQTVV Y NAGMGVGV ...String: RVRSITLGNS TITTQECANV VVGYGVWPSY LQDNEATAED QPTQPDVATC RFYTLDSIQW QKESDGWWWK FPEALKNMGL FGQNMEYHY LGRSGYTIHV QCNASKFHQG CLLVVCVPEA EMGCSDVERE VVAASLSSED TAKSFSRTES NGQHTVQTVV Y NAGMGVGV GNLTIFPHQW INLRTNNSAT IVMPYINSVP MDNMFRHYNF TLMIIPFAKL EYTEQASNYV PITVTVAPMC AE YNGLRLA SHQ UniProtKB: Genome polyprotein |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E3 Echovirus E3 |
| Molecular weight | Theoretical: 25.896449 KDa |
| Sequence | String: MLTPGSNQFL TSDDFQSPSA MPQFDVTPEM KIPGEVHNLM EIAEVDSVVP VNNTKENINS MEAYRIPVTG GDQLHTQVFG FQMQPGLNS VFKRTLLGEI LNYYAHWSGS VKLTFVFCGS AMATGKFLLA YSPPGASPPQ NRKQAMLGTH VIWDVGLQSS C VLCIPWIS ...String: MLTPGSNQFL TSDDFQSPSA MPQFDVTPEM KIPGEVHNLM EIAEVDSVVP VNNTKENINS MEAYRIPVTG GDQLHTQVFG FQMQPGLNS VFKRTLLGEI LNYYAHWSGS VKLTFVFCGS AMATGKFLLA YSPPGASPPQ NRKQAMLGTH VIWDVGLQSS C VLCIPWIS QTHYRLVQQD EYTSAGYVTC WYQTGLIVPP GAPPSCTILC FASACNDFSV RNLRDTPFIE QTQLLQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Heavy chain of 6D10
| Macromolecule | Name: Heavy chain of 6D10 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.9382 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQQSGPE LARPWASVKI SCQAFYTFSN YGMQWVKQSP GQGLEWIGPF YPGNADTSYN QKFKGKATLT ADKSSSTAYM QFSSLTSED SAVYYCARVV ATTDFDYWGQ GTTVTVSS |
-Macromolecule #5: Light chain of 6D10
| Macromolecule | Name: Light chain of 6D10 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.652158 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YIEASQSPSS LAVSVGEKVT MSCKSSQSLL YSSNQKNYLA WFQQKPGQSP KLLIYWASTR ESGVPDRFTG SGSGTDFTLT ISSVKAEDL AVYYCQQYYS YPYTFGGGTK LKIKR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)