+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of pentameric MotA from Aquifex aeolicus | ||||||||||||||||||||||||
 Map data Map data | MotA 5mer postprocessed density map from dataset1 | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Bacterial flagellum / stator protein / Aquifex aeolicus / single particle Cryo-EM / MotA / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum-dependent swarming motility / proton transmembrane transport / chemotaxis / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Aquifex aeolicus (bacteria) / Aquifex aeolicus (bacteria) /   Aquifex aeolicus VF5 (bacteria) Aquifex aeolicus VF5 (bacteria) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||||||||
 Authors Authors | Nishikino T / Takekawa N / Kishikawa J / Hirose M / Onoe S / Kato T / Imada K | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2022 Journal: Biochem Biophys Res Commun / Year: 2022Title: Structure of MotA, a flagellar stator protein, from hyperthermophile. Authors: Tatsuro Nishikino / Norihiro Takekawa / Duy Phuoc Tran / Jun-Ichi Kishikawa / Mika Hirose / Sakura Onoe / Seiji Kojima / Michio Homma / Akio Kitao / Takayuki Kato / Katsumi Imada /  Abstract: Many motile bacteria swim and swarm toward favorable environments using the flagellum, which is rotated by a motor embedded in the inner membrane. The motor is composed of the rotor and the stator, ...Many motile bacteria swim and swarm toward favorable environments using the flagellum, which is rotated by a motor embedded in the inner membrane. The motor is composed of the rotor and the stator, and the motor torque is generated by the change of the interaction between the rotor and the stator induced by the ion flow through the stator. A stator unit consists of two types of membrane proteins termed A and B. Recent cryo-EM studies on the stators from mesophiles revealed that the stator consists of five A and two B subunits, whereas the low-resolution EM analysis showed that purified hyperthermophilic MotA forms a tetramer. To clarify the assembly formation and factors enhancing thermostability of the hyperthermophilic stator, we determined the cryo-EM structure of MotA from Aquifex aeolicus (Aa-MotA), a hyperthermophilic bacterium, at 3.42 Å resolution. Aa-MotA forms a pentamer with pseudo C5 symmetry. A simulated model of the Aa-MotAMotB stator complex resembles the structures of mesophilic stator complexes, suggesting that Aa-MotA can assemble into a pentamer equivalent to the stator complex without MotB. The distribution of hydrophobic residues of MotA pentamers suggests that the extremely hydrophobic nature in the subunit boundary and the transmembrane region is a key factor to stabilize hyperthermophilic Aa-MotA. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34203.map.gz emd_34203.map.gz | 96.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34203-v30.xml emd-34203-v30.xml emd-34203.xml emd-34203.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34203_fsc.xml emd_34203_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_34203.png emd_34203.png | 85.2 KB | ||
| Masks |  emd_34203_msk_1.map emd_34203_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34203.cif.gz emd-34203.cif.gz | 6.3 KB | ||
| Others |  emd_34203_half_map_1.map.gz emd_34203_half_map_1.map.gz emd_34203_half_map_2.map.gz emd_34203_half_map_2.map.gz | 81 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34203 http://ftp.pdbj.org/pub/emdb/structures/EMD-34203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34203 | HTTPS FTP |
-Validation report
| Summary document |  emd_34203_validation.pdf.gz emd_34203_validation.pdf.gz | 1019.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34203_full_validation.pdf.gz emd_34203_full_validation.pdf.gz | 1019.2 KB | Display | |
| Data in XML |  emd_34203_validation.xml.gz emd_34203_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_34203_validation.cif.gz emd_34203_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34203 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34203 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34203 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34203 | HTTPS FTP |
-Related structure data
| Related structure data |  8gqyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34203.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34203.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MotA 5mer postprocessed density map from dataset1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||
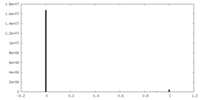
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34203_msk_1.map emd_34203_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
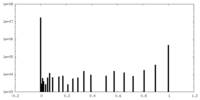
| Density Histograms |
-Half map: half map 2 from dataset 1
| File | emd_34203_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 from dataset 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
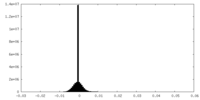
| Density Histograms |
-Half map: half map 1 from dataset 1
| File | emd_34203_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 from dataset 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
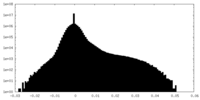
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric MotA from Aquifex aeolicus
| Entire | Name: Pentameric MotA from Aquifex aeolicus |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric MotA from Aquifex aeolicus
| Supramolecule | Name: Pentameric MotA from Aquifex aeolicus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Motility protein A
| Macromolecule | Name: Motility protein A / type: protein_or_peptide / ID: 1 Details: Met1 to His6 is translation enhancing element sequence. 6 His residues on C-terminal are purification tag. Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus VF5 (bacteria) / Strain: VF5 Aquifex aeolicus VF5 (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 28.863195 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNHKVHMDVG TIIGIIAAFL LILISILIGG SITAFINVPS IFIVVGGGMA AAMGAFPLKD FIRGVLAIKK AFLWKPPDLN DVIETIGEI ASKVRKEGIL ALEGDIELYY QKDPLLGDMI RMLVDGIDIN DIKATAEMAL AQLDEKMSTE VAVWEKLADL F PAFGMIGT ...String: MNHKVHMDVG TIIGIIAAFL LILISILIGG SITAFINVPS IFIVVGGGMA AAMGAFPLKD FIRGVLAIKK AFLWKPPDLN DVIETIGEI ASKVRKEGIL ALEGDIELYY QKDPLLGDMI RMLVDGIDIN DIKATAEMAL AQLDEKMSTE VAVWEKLADL F PAFGMIGT LIGLIQMLRN LNDPSALGPG MAVALITTLY GAILANAFAI PVANKLKKAK DMEVLVKTIY IEAIEKIQKG EN PNVVKQE AAIMLGVELP EEVHHHHHH UniProtKB: Motility protein A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 6299 / Average exposure time: 2.22 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 60.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)