+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Designed pH-responsive P22 VLP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P22 coat protein / designed protein / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Major capsid protein Gp5 / P22 coat protein - gene protein 5 / viral procapsid / viral procapsid maturation / T=7 icosahedral viral capsid / viral capsid / identical protein binding / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Lederbergvirus Lederbergvirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.02 Å | |||||||||
 Authors Authors | Kim KJ / Kim G / Bae JH / Song JJ / Kim HS | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Adv Healthc Mater / Year: 2024 Journal: Adv Healthc Mater / Year: 2024Title: A pH-Responsive Virus-Like Particle as a Protein Cage for a Targeted Delivery. Authors: Kwan-Jip Kim / Gijeong Kim / Jin-Ho Bae / Ji-Joon Song / Hak-Sung Kim /  Abstract: A stimuli-responsive protein self-assembly offers promising utility as a protein nanocage for biotechnological and medical applications. Herein, the development of a virus-like particle (VLP) that ...A stimuli-responsive protein self-assembly offers promising utility as a protein nanocage for biotechnological and medical applications. Herein, the development of a virus-like particle (VLP) that undergoes a transition between assembly and disassembly under a neutral and acidic pH, respectively, for a targeted delivery is reported. The structure of the bacteriophage P22 coat protein is used for the computational design of coat subunits that self-assemble into a pH-responsive VLP. Subunit designs are generated through iterative computational cycles of histidine substitutions and evaluation of the interaction energies among the subunits under an acidic and neutral pH. The top subunit designs are tested and one that is assembled into a VLP showing the highest pH-dependent structural transition is selected. The cryo-EM structure of the VLP is determined, and the structural basis of a pH-triggered disassembly is delineated. The utility of the designed VLP is exemplified through the targeted delivery of a cytotoxic protein cargo into tumor cells in a pH-dependent manner. These results provide strategies for the development of self-assembling protein architectures with new functionality for diverse applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34155.map.gz emd_34155.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34155-v30.xml emd-34155-v30.xml emd-34155.xml emd-34155.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34155.png emd_34155.png | 108 KB | ||
| Filedesc metadata |  emd-34155.cif.gz emd-34155.cif.gz | 5.8 KB | ||
| Others |  emd_34155_half_map_1.map.gz emd_34155_half_map_1.map.gz emd_34155_half_map_2.map.gz emd_34155_half_map_2.map.gz | 11.7 MB 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34155 http://ftp.pdbj.org/pub/emdb/structures/EMD-34155 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34155 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34155 | HTTPS FTP |
-Related structure data
| Related structure data |  8gn5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34155.map.gz / Format: CCP4 / Size: 12.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34155.map.gz / Format: CCP4 / Size: 12.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2995 Å | ||||||||||||||||||||||||||||||||||||
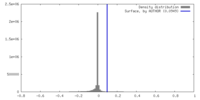
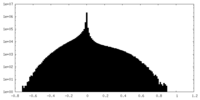
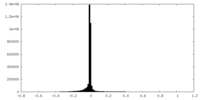
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34155_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
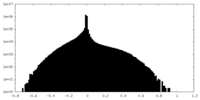
| Density Histograms |
-Half map: #1
| File | emd_34155_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lederbergvirus
| Entire | Name:  Lederbergvirus Lederbergvirus |
|---|---|
| Components |
|
-Supramolecule #1: Lederbergvirus
| Supramolecule | Name: Lederbergvirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 186794 / Sci species name: Lederbergvirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Lederbergvirus Lederbergvirus |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lederbergvirus Lederbergvirus |
| Molecular weight | Theoretical: 46.40118 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NEGQIVTLAV DEIIETISAI TPMAQKAKKY TPPAASMQRH SNTIWMPVEQ HSPTQEGWDL TDKATGLLEL NVAVNMGEPD NDFFQLRAD DLRDETAYRR RIQSAARKLA NNVELKVANM AAEMGSLVIT SPDAIGTNTA DAWNFVADAE HIMFSRELNR D MGTSYFFN ...String: NEGQIVTLAV DEIIETISAI TPMAQKAKKY TPPAASMQRH SNTIWMPVEQ HSPTQEGWDL TDKATGLLEL NVAVNMGEPD NDFFQLRAD DLRDETAYRR RIQSAARKLA NNVELKVANM AAEMGSLVIT SPDAIGTNTA DAWNFVADAE HIMFSRELNR D MGTSYFFN PQDYKKAGYD LTKRDIFGRI PEEAYRDGTI QRQVAGFDDV LRSPKLPVLT KSTATGITVS GAQSFKPVAW QL DNDGNKV NVDNRFATVT LSATTGMKRG DKISFAGVKF LGQMAKHVLA QDATFSVVRV VDGTHVEITP KPVALDDVSL SPE QRAYAN VNTSLADAMA VNILNVKDAR TNVFWADDAI RIVSQPIPAN HELFAGMKTT SFSIPDVGLN GIFATQGDIS TLSG LCRIA LWYGVNATRP EAIGVGLPGQ UniProtKB: Major capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 17 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: PBS buffer pH 7.4 | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: The grid was glow discharged at 15 mA for 1 min. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1654 / Average exposure time: 52.69 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 92000 |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)