+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a mutated membrane-bound glycosyltransferase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationfungal-type cell wall polysaccharide biosynthetic process / 1,3-beta-glucan synthase / 1,3-beta-D-glucan synthase activity / (1->3)-beta-D-glucan biosynthetic process / 1,3-beta-D-glucan synthase complex / fungal-type cell wall biogenesis / cellular bud / ascospore wall assembly / actin cortical patch / cellular bud tip ...fungal-type cell wall polysaccharide biosynthetic process / 1,3-beta-glucan synthase / 1,3-beta-D-glucan synthase activity / (1->3)-beta-D-glucan biosynthetic process / 1,3-beta-D-glucan synthase complex / fungal-type cell wall biogenesis / cellular bud / ascospore wall assembly / actin cortical patch / cellular bud tip / fungal-type cell wall / cellular bud neck / regulation of cell size / positive regulation of endocytosis / cell periphery / mitochondrion / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
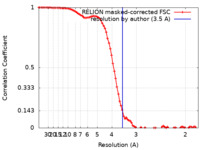
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Hu XL / Yang P / Zhang M / Liu XT / Yu HJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural and mechanistic insights into fungal β-1,3-glucan synthase FKS1. Authors: Xinlin Hu / Ping Yang / Changdong Chai / Jia Liu / Huanhuan Sun / Yanan Wu / Mingjie Zhang / Min Zhang / Xiaotian Liu / Hongjun Yu /  Abstract: The membrane-integrated synthase FKS is involved in the biosynthesis of β-1,3-glucan, the core component of the fungal cell wall. FKS is the target of widely prescribed antifungal drugs, including ...The membrane-integrated synthase FKS is involved in the biosynthesis of β-1,3-glucan, the core component of the fungal cell wall. FKS is the target of widely prescribed antifungal drugs, including echinocandin and ibrexafungerp. Unfortunately, the mechanism of action of FKS remains enigmatic and this has hampered development of more effective medicines targeting the enzyme. Here we present the cryo-electron microscopy structures of Saccharomyces cerevisiae FKS1 and the echinocandin-resistant mutant FKS1(S643P). These structures reveal the active site of the enzyme at the membrane-cytoplasm interface and a glucan translocation path spanning the membrane bilayer. Multiple bound lipids and notable membrane distortions are observed in the FKS1 structures, suggesting active FKS1-membrane interactions. Echinocandin-resistant mutations are clustered at a region near TM5-6 and TM8 of FKS1. The structure of FKS1(S643P) reveals altered lipid arrangements in this region, suggesting a drug-resistant mechanism of the mutant enzyme. The structures, the catalytic mechanism and the molecular insights into drug-resistant mutations of FKS1 revealed in this study advance the mechanistic understanding of fungal β-1,3-glucan biosynthesis and establish a foundation for developing new antifungal drugs by targeting FKS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34115.map.gz emd_34115.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34115-v30.xml emd-34115-v30.xml emd-34115.xml emd-34115.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34115_fsc.xml emd_34115_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34115.png emd_34115.png | 62.3 KB | ||
| Others |  emd_34115_half_map_1.map.gz emd_34115_half_map_1.map.gz emd_34115_half_map_2.map.gz emd_34115_half_map_2.map.gz | 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34115 http://ftp.pdbj.org/pub/emdb/structures/EMD-34115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34115 | HTTPS FTP |
-Validation report
| Summary document |  emd_34115_validation.pdf.gz emd_34115_validation.pdf.gz | 939.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34115_full_validation.pdf.gz emd_34115_full_validation.pdf.gz | 939.3 KB | Display | |
| Data in XML |  emd_34115_validation.xml.gz emd_34115_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_34115_validation.cif.gz emd_34115_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34115 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34115 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34115 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34115 | HTTPS FTP |
-Related structure data
| Related structure data |  7yuyMC  7xe4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34115.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34115.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34115_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
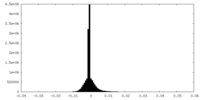
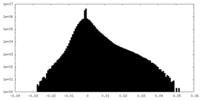
| Density Histograms |
-Half map: #2
| File | emd_34115_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
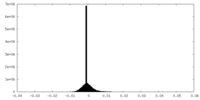
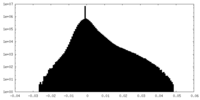
| Density Histograms |
- Sample components
Sample components
-Entire : membrane-bound glycosyltransferase
| Entire | Name: membrane-bound glycosyltransferase |
|---|---|
| Components |
|
-Supramolecule #1: membrane-bound glycosyltransferase
| Supramolecule | Name: membrane-bound glycosyltransferase / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 215 KDa |
-Macromolecule #1: 1,3-beta-glucan synthase component FKS1
| Macromolecule | Name: 1,3-beta-glucan synthase component FKS1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: 1,3-beta-glucan synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 215.086203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTDQQPYQG QTDYTQGPGN GQSQEQDYDQ YGQPLYPSQA DGYYDPNVAA GTEADMYGQQ PPNESYDQDY TNGEYYGQPP NMAAQDGEN FSDFSSYGPP GTPGYDSYGG QYTASQMSYG EPNSSGTSTP IYGNYDPNAI AMALPNEPYP AWTADSQSPV S IEQIEDIF ...String: MNTDQQPYQG QTDYTQGPGN GQSQEQDYDQ YGQPLYPSQA DGYYDPNVAA GTEADMYGQQ PPNESYDQDY TNGEYYGQPP NMAAQDGEN FSDFSSYGPP GTPGYDSYGG QYTASQMSYG EPNSSGTSTP IYGNYDPNAI AMALPNEPYP AWTADSQSPV S IEQIEDIF IDLTNRLGFQ RDSMRNMFDH FMVLLDSRSS RMSPDQALLS LHADYIGGDT ANYKKWYFAA QLDMDDEIGF RN MSLGKLS RKARKAKKKN KKAMEEANPE DTEETLNKIE GDNSLEAADF RWKAKMNQLS PLERVRHIAL YLLCWGEANQ VRF TAECLC FIYKCALDYL DSPLCQQRQE PMPEGDFLNR VITPIYHFIR NQVYEIVDGR FVKRERDHNK IVGYDDLNQL FWYP EGIAK IVLEDGTKLI ELPLEERYLR LGDVVWDDVF FKTYKETRTW LHLVTNFNRI WVMHISIFWM YFAYNSPTFY THNYQ QLVD NQPLAAYKWA SCALGGTVAS LIQIVATLCE WSFVPRKWAG AQHLSRRFWF LCIIFGINLG PIIFVFAYDK DTVYST AAH VVAAVMFFVA VATIIFFSIM PLGGLFTSYM KKSTRRYVAS QTFTAAFAPL HGLDRWMSYL VWVTVFAAKY SESYYFL VL PLRDPIRILS TTAMRCTGEY WWGAVLCKVQ PKIVLGLVIA TDFILFFLDT YLWYIIVNTI FSVGKSFYLG ISILTPWR N IFTRLPKRIY SKILATTDME IKYKPKVLIS QVWNAIIISM YREHLLAIDH VQKLLYHQVP SEIEGKRTLR APTFFVSQD DNNFETEFFP RDSEAERRIS FFAQSLSTPI PEPLPVDNMP TFTVLTPHYA ERILLSLREI IREDDQFSRV TLLEYLKQLH PVEWECFVK DTKILAEETA AYEGNENEAE KEDALKSQID DLPFYCIGFK SAAPEYTLRT RIWASLRSQT LYRTISGFMN Y SRAIKLLY RVENPEIVQM FGGNAEGLER ELEKMARRKF KFLVSMQRLA KFKPHELENA EFLLRAYPDL QIAYLDEEPP LT EGEEPRI YSALIDGHCE ILDNGRRRPK FRVQLSGNPI LGDGKSDNQN HALIFYRGEY IQLIDANQDN YLEECLKIRS VLA EFEELN VEQVNPYAPG LRYEEQTTNH PVAIVGAREY IFSENSGVLG DVAAGKEQTF GTLFARTLSQ IGGKLHYGHP DFIN ATFMT TRGGVSKAQK GLHLNEDIYA GMNAMLRGGR IKHCEYYQCG KGRDLGFGTI LNFTTKIGAG MGEQMLSREY YYLGT QLPV DRFLTFYYAH PGFHLNNLFI QLSLQMFMLT LVNLSSLAHE SIMCIYDRNK PKTDVLVPIG CYNFQPAVDW VRRYTL SIF IVFWIAFVPI VVQELIERGL WKATQRFFCH LLSLSPMFEV FAGQIYSSAL LSDLAIGGAR YISTGRGFAT SRIPFSI LY SRFAGSAIYM GARSMLMLLF GTVAHWQAPL LWFWASLSSL IFAPFVFNPH QFAWEDFFLD YRDYIRWLSR GNNQYHRN S WIGYVRMSRA RITGFKRKLV GDESEKAAGD ASRAHRTNLI MAEIIPCAIY AAGCFIAFTF INAQTGVKTT DDDRVNSVL RIIICTLAPI AVNLGVLFFC MGMSCCSGPL FGMCCKKTGS VMAGIAHGVA VIVHIAFFIV MWVLESFNFV RMLIGVVTCI QCQRLIFHC MTALMLTREF KNDHANTAFW TGKWYGKGMG YMAWTQPSRE LTAKVIELSE FAADFVLGHV ILICQLPLII I PKIDKFHS IMLFWLKPSR QIRPPIYSLK QTRLRKRMVK KYCSLYFLVL AIFAGCIIGP AVASAKIHKH IGDSLDGVVH NL FQPINTT NNDTGSQMST YQSHYYTHTP SLKTWSTIK |
-Macromolecule #4: nonane
| Macromolecule | Name: nonane / type: ligand / ID: 4 / Number of copies: 7 / Formula: DD9 |
|---|---|
| Molecular weight | Theoretical: 128.255 Da |
| Chemical component information | 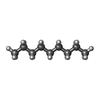 ChemComp-DD9: |
-Macromolecule #5: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 5 / Number of copies: 6 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #6: TETRADECANE
| Macromolecule | Name: TETRADECANE / type: ligand / ID: 6 / Number of copies: 1 / Formula: C14 |
|---|---|
| Molecular weight | Theoretical: 198.388 Da |
| Chemical component information |  ChemComp-C14: |
-Macromolecule #7: HEPTANE
| Macromolecule | Name: HEPTANE / type: ligand / ID: 7 / Number of copies: 7 / Formula: HP6 |
|---|---|
| Molecular weight | Theoretical: 100.202 Da |
| Chemical component information |  ChemComp-HP6: |
-Macromolecule #8: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 8 / Number of copies: 3 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #9: (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~...
| Macromolecule | Name: (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate type: ligand / ID: 9 / Number of copies: 1 / Formula: XKP |
|---|---|
| Molecular weight | Theoretical: 495.587 Da |
| Chemical component information | 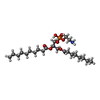 ChemComp-XKP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X