+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of WTAP-VIRMA in the m6A writer complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA N6-methyladenosine methyltransferase complex / mRNA alternative polyadenylation / : / regulation of alternative mRNA splicing, via spliceosome / Processing of Capped Intron-Containing Pre-mRNA / RNA splicing / mRNA processing / nuclear membrane / nuclear body / nuclear speck ...RNA N6-methyladenosine methyltransferase complex / mRNA alternative polyadenylation / : / regulation of alternative mRNA splicing, via spliceosome / Processing of Capped Intron-Containing Pre-mRNA / RNA splicing / mRNA processing / nuclear membrane / nuclear body / nuclear speck / cell cycle / RNA binding / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Yan XH / Guan ZY / Tang C / Yin P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: AI-empowered integrative structural characterization of mA methyltransferase complex. Authors: Xuhui Yan / Kai Pei / Zeyuan Guan / Feiqing Liu / Junjun Yan / Xiaohuan Jin / Qiang Wang / Mengjun Hou / Chun Tang / Ping Yin /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33807.map.gz emd_33807.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33807-v30.xml emd-33807-v30.xml emd-33807.xml emd-33807.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33807.png emd_33807.png | 45.1 KB | ||
| Filedesc metadata |  emd-33807.cif.gz emd-33807.cif.gz | 5.9 KB | ||
| Others |  emd_33807_half_map_1.map.gz emd_33807_half_map_1.map.gz emd_33807_half_map_2.map.gz emd_33807_half_map_2.map.gz | 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33807 http://ftp.pdbj.org/pub/emdb/structures/EMD-33807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33807 | HTTPS FTP |
-Validation report
| Summary document |  emd_33807_validation.pdf.gz emd_33807_validation.pdf.gz | 911.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33807_full_validation.pdf.gz emd_33807_full_validation.pdf.gz | 911.5 KB | Display | |
| Data in XML |  emd_33807_validation.xml.gz emd_33807_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_33807_validation.cif.gz emd_33807_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33807 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33807 | HTTPS FTP |
-Related structure data
| Related structure data |  7yg4MC  7yfjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33807.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33807.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33807_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
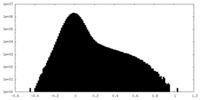
| Projections & Slices |
| ||||||||||||
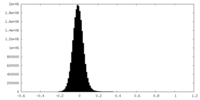
| Density Histograms |
-Half map: #2
| File | emd_33807_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
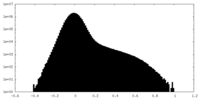
| Projections & Slices |
| ||||||||||||
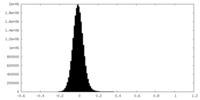
| Density Histograms |
- Sample components
Sample components
-Entire : WTAP-VIRMA complex
| Entire | Name: WTAP-VIRMA complex |
|---|---|
| Components |
|
-Supramolecule #1: WTAP-VIRMA complex
| Supramolecule | Name: WTAP-VIRMA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein virilizer homolog
| Macromolecule | Name: Protein virilizer homolog / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 123.716016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASVKLTELL DLYREDRGAK WVTALEEIPS LIIKGLSYLQ LKNTKQDSLG QLVDWTMQAL NLQVALRQPI ALNVRQLKAG TKLVSSLAE CGAQGVTGLL QAGVISGLFE LLFADHVSSS LKLNAFKALD SVISMTEGME AFLRGRQNEK SGYQKLLELI L LDQTVRVV ...String: MASVKLTELL DLYREDRGAK WVTALEEIPS LIIKGLSYLQ LKNTKQDSLG QLVDWTMQAL NLQVALRQPI ALNVRQLKAG TKLVSSLAE CGAQGVTGLL QAGVISGLFE LLFADHVSSS LKLNAFKALD SVISMTEGME AFLRGRQNEK SGYQKLLELI L LDQTVRVV TAGSAILQKC HFYEVLSEIK RLGDHLAEKT SSLPNHSEPD HDTDAGLERT NPEYENEVEA SMDMDLLESS NI SEGEIER LINLLEEVFH LMETAPHTMI QQPVKSFPTM ARITGPPERD DPYPVLFRYL HSHHFLELVT LLLSIPVTSA HPG VLQATK DVLKFLAQSQ KGLLFFMSEY EATNLLIRAL CHFYDQDEEE GLQSDGVIDD AFALWLQDST QTLQCITELF SHFQ RCTAS EETDHSDLLG TLHNLYLITF NPVGRSAVGH VFSLEKNLQS LITLMEYYSK EALGDSKSKK SVAYNYACIL ILVVV QSSS DVQMLEQHAA SLLKLCKADE NNAKLQELGK WLEPLKNLRF EINCIPNLIE YVKQNIDNLM TPEGVGLTTA LRVLCN VAC PPPPVEGQQK DLKWNLAVIQ LFSAEGMDTF IRVLQKLNSI LTQPWRLHVN MGTTLHRVTT ISMARCTLTL LKTMLTE LL RGGSFEFKDM RVPSALVTLH MLLCSIPLSG RLDSDEQKIQ NDIIDILLTF TQGVNEKLTI SEETLANNTW SLMLKEVL S SILKVPEGFF SGLILLSELL PLPLPMQTTQ VIEPHDISVA LNTRKLWSMH LHVQAKLLQE IVRSFSGTTC QPIQHMLRR ICVQLCDLAS PTALLIMRTV LDLIVEDLQS TSEDKEKQYT SQTTRLLALL DALASHKACK LAILHLINGT IKGDERYAEI FQDLLALVR SPGDSVIRQQ CVEYVTSILQ SLCDQDIALI LPSSSEGSIS ELEQLSNSLP NKELMTSICD CLLATLANSE S SYNCLLTC VRTMMFLAEH DYGLFHLKSS LRKNSSALHS LLKRVVSTFS KDTGELASSF LEFMRQILNS DTIGCCGDDN GL MEVEGAH TSRTMSINAA ELKQLLQSKE ESPENLFLEL EKLVLEHSKD DDNLDSLLDS VVGLKQMLES UniProtKB: Protein virilizer homolog |
-Macromolecule #2: Pre-mRNA-splicing regulator WTAP
| Macromolecule | Name: Pre-mRNA-splicing regulator WTAP / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.107188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HSGDEVDAGS GHMTNEEPLP KKVRLSETDF KVMARDELIL RWKQYEAYVQ ALEGKYTDLN SNDVTGLRES EEKLKQQQQ ESARRENILV MRLATKEQEM QECTTQIQYL KQVQQPSVAQ LRSTMVDPAI NLFFLKMKGE LEQTKDKLEQ A QNELSAWK ...String: MHHHHHHHHH HSGDEVDAGS GHMTNEEPLP KKVRLSETDF KVMARDELIL RWKQYEAYVQ ALEGKYTDLN SNDVTGLRES EEKLKQQQQ ESARRENILV MRLATKEQEM QECTTQIQYL KQVQQPSVAQ LRSTMVDPAI NLFFLKMKGE LEQTKDKLEQ A QNELSAWK FTPDSQTGKK LMAKCRMLIQ ENQELGRQLS QGRIAQLEAE LALQKKYSEE LKSSQDELND FIIQLDEEVE GM QSTILVL QQQLKETRQQ LAQYQQQQSQ ASAPSTSRTT ASEPVEQSEA TSKDCSRL UniProtKB: Pre-mRNA-splicing regulator WTAP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X