+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | malaria / nucleoside transporter / GSK4 / TRANSPORT PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTransport of nucleosides and free purine and pyrimidine bases across the plasma membrane / Ribavirin ADME / adenosine transport / Azathioprine ADME / nucleoside transmembrane transporter activity / purine nucleobase transport / bioluminescence / generation of precursor metabolites and energy / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
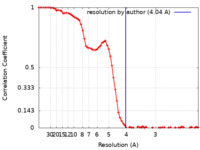
| Method | single particle reconstruction / cryo EM / Resolution: 4.04 Å | |||||||||
 Authors Authors | Wang C / Yu LY / Li JL / Ren RB / Deng D | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1. Authors: Chen Wang / Leiye Yu / Jiying Zhang / Yanxia Zhou / Bo Sun / Qingjie Xiao / Minhua Zhang / Huayi Liu / Jinhong Li / Jialu Li / Yunzi Luo / Jie Xu / Zhong Lian / Jingwen Lin / Xiang Wang / ...Authors: Chen Wang / Leiye Yu / Jiying Zhang / Yanxia Zhou / Bo Sun / Qingjie Xiao / Minhua Zhang / Huayi Liu / Jinhong Li / Jialu Li / Yunzi Luo / Jie Xu / Zhong Lian / Jingwen Lin / Xiang Wang / Peng Zhang / Li Guo / Ruobing Ren / Dong Deng /  Abstract: By lacking de novo purine biosynthesis enzymes, Plasmodium falciparum requires purine nucleoside uptake from host cells. The indispensable nucleoside transporter ENT1 of P. falciparum facilitates ...By lacking de novo purine biosynthesis enzymes, Plasmodium falciparum requires purine nucleoside uptake from host cells. The indispensable nucleoside transporter ENT1 of P. falciparum facilitates nucleoside uptake in the asexual blood stage. Specific inhibitors of PfENT1 prevent the proliferation of P. falciparum at submicromolar concentrations. However, the substrate recognition and inhibitory mechanism of PfENT1 are still elusive. Here, we report cryo-EM structures of PfENT1 in apo, inosine-bound, and inhibitor-bound states. Together with in vitro binding and uptake assays, we identify that inosine is the primary substrate of PfENT1 and that the inosine-binding site is located in the central cavity of PfENT1. The endofacial inhibitor GSK4 occupies the orthosteric site of PfENT1 and explores the allosteric site to block the conformational change of PfENT1. Furthermore, we propose a general "rocker switch" alternating access cycle for ENT transporters. Understanding the substrate recognition and inhibitory mechanisms of PfENT1 will greatly facilitate future efforts in the rational design of antimalarial drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33756.map.gz emd_33756.map.gz | 27.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33756-v30.xml emd-33756-v30.xml emd-33756.xml emd-33756.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33756_fsc.xml emd_33756_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_33756.png emd_33756.png | 33.1 KB | ||
| Filedesc metadata |  emd-33756.cif.gz emd-33756.cif.gz | 6.1 KB | ||
| Others |  emd_33756_half_map_1.map.gz emd_33756_half_map_1.map.gz emd_33756_half_map_2.map.gz emd_33756_half_map_2.map.gz | 26.6 MB 26.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33756 http://ftp.pdbj.org/pub/emdb/structures/EMD-33756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33756 | HTTPS FTP |
-Validation report
| Summary document |  emd_33756_validation.pdf.gz emd_33756_validation.pdf.gz | 770.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33756_full_validation.pdf.gz emd_33756_full_validation.pdf.gz | 770.4 KB | Display | |
| Data in XML |  emd_33756_validation.xml.gz emd_33756_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_33756_validation.cif.gz emd_33756_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33756 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33756 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33756 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33756 | HTTPS FTP |
-Related structure data
| Related structure data |  7ydqMC  7wn0C  7wn1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33756.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33756.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33756_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
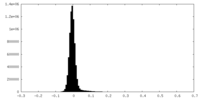
| Density Histograms |
-Half map: #2
| File | emd_33756_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
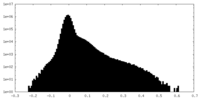
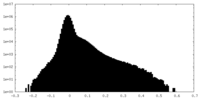
| Density Histograms |
- Sample components
Sample components
-Entire : nucleoside/nucleobase transporter fusion with GFP
| Entire | Name: nucleoside/nucleobase transporter fusion with GFP |
|---|---|
| Components |
|
-Supramolecule #1: nucleoside/nucleobase transporter fusion with GFP
| Supramolecule | Name: nucleoside/nucleobase transporter fusion with GFP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: the cDNA of the GFP was cloned into PfNT1 between K370 and K371 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.5 kDa/nm |
-Macromolecule #1: Nucleoside transporter 1,Green fluorescent protein
| Macromolecule | Name: Nucleoside transporter 1,Green fluorescent protein / type: protein_or_peptide / ID: 1 Details: PfNT1(Y190A) fused with GFP the GFP was inserted into PfNT1 between K370 and K371 Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.035766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HSGDEVDAGS GHMSTGKESS KAYADIESRG DYKDDGKKGS TLSSKQHFML SLTFILIGLS SLNVWNTALG LNINFKYNT FQITGLVCSS IVALFVEIPK IMLPFLLGGL SILCAGFQIS HSFFTDTQFD TYCLVAFIVI GVVAGLAQTI A FNIGSTME ...String: MHHHHHHHHH HSGDEVDAGS GHMSTGKESS KAYADIESRG DYKDDGKKGS TLSSKQHFML SLTFILIGLS SLNVWNTALG LNINFKYNT FQITGLVCSS IVALFVEIPK IMLPFLLGGL SILCAGFQIS HSFFTDTQFD TYCLVAFIVI GVVAGLAQTI A FNIGSTME DNMGGYMSAG IGISGVFIFV INLLLDQFVS PEKHYGVNKA KLLALYIICE LCLILAIVFC VCNLDLTNKN NK KDEENKE NNATLSYMEL FKDSYKAILT MFLVNWLTLQ LFPGVGHKKW QESHNISDYN VTIIVGMFQV FDFLSRYPPN LTH IKIFKN FTFSLNKLLV ANSLRLLFIP WFILNACVDH PFFKNIVQQC VCMAMLAFTN GWFNTVPFLV FVKELKMVSK GEEL FTGVV PILVELDGDV NGHKFSVSGE GEGDATYGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKQHD FFKSA MPEG YVQERTIFFK DDGNYKTRAE VKFEGDTLVN RIELKGIDFK EDGNILGHKL EYNYNSHNVY IMADKQKNGI KANFKI RHN IEDGSVQLAD HYQQNTPIGD GPVLLPDNHY LSTQSALSKD PNEKRDHMVL LEFVTAAGIT LGMDELYKKA KKKKEIE II STFLVIAMFV GLFCGIWTTY IYNLFNIVLP KPDLPPIDVT Q UniProtKB: Nucleoside transporter 1, Green fluorescent protein, Nucleoside transporter 1 |
-Macromolecule #2: 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxa...
| Macromolecule | Name: 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide type: ligand / ID: 2 / Number of copies: 1 / Formula: IRX |
|---|---|
| Molecular weight | Theoretical: 341.404 Da |
| Chemical component information | 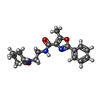 ChemComp-IRX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 52.452 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X