+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of Echo 18 and FcRn at pH7.4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Receptor / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationIgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor / IgG binding / beta-2-microglobulin binding / endosome membrane / immune response / external side of plasma membrane / extracellular space Similarity search - Function | |||||||||
| Biological species |  Echovirus E18 / Echovirus E18 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
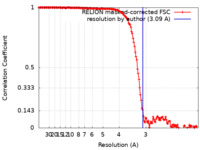
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Liu CC / Qu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2022 Journal: mBio / Year: 2022Title: Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18. Authors: Xiangpeng Chen / Xiao Qu / Congcong Liu / Yong Zhang / Guigen Zhang / Pu Han / Yali Duan / Qi Li / Liang Wang / Wenjing Ruan / Peiyi Wang / Wensheng Wei / George F Gao / Xin Zhao / Zhengde Xie /  Abstract: Virus-receptor interactions determine viral host range and tissue tropism. CD55 and human neonatal Fc receptor (FcRn) were found to be the binding and uncoating receptors for some of the echovirus- ...Virus-receptor interactions determine viral host range and tissue tropism. CD55 and human neonatal Fc receptor (FcRn) were found to be the binding and uncoating receptors for some of the echovirus-related enterovirus species B serotypes in our previous study. Echovirus 18 (E18), as a member of enterovirus species B, is a significant causative agent of aseptic meningitis and viral encephalitis in children. However, it does not use CD55 as a critical host factor. We conducted CRISPR/Cas9 knockout screening to determine the receptors and entry mechanisms and identified FcRn working as a dual-function receptor for E18. Knockout of and , which encode the two subunits of FcRn, prevented infection by E18 and other echoviruses in the same physiological cluster. We then elucidated the underlying molecular mechanism of receptor recognition by E18 using cryogenic electron microscopy. The binding of the FCGRT subunit to the canyon region rotates the residues around the pocket, triggering the release of the pocket factor as observed for other enterovirus species B members. E18 is a member of enterovirus species B. As one of the most common enterovirus serotypes in nonpolio enterovirus detection, it easily infects children and causes various clinical symptoms. Aseptic meningitis and viral encephalitis are the most commonly reported syndromes associated with E18. No effective antiviral drugs or approved vaccines are available. Previous studies showed that CD55 and FcRn were the binding and uncoating receptors for some echoviruses. However, we found that CD55 is not the critical host factor for E18. Thus, we want to determine the receptors and elucidate the entry mechanism of E18. Our findings reveal that FcRn is a two-in-one attachment-uncoating receptor for E18. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33499.map.gz emd_33499.map.gz | 98.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33499-v30.xml emd-33499-v30.xml emd-33499.xml emd-33499.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33499_fsc.xml emd_33499_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_33499.png emd_33499.png | 150.3 KB | ||
| Masks |  emd_33499_msk_1.map emd_33499_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33499.cif.gz emd-33499.cif.gz | 7.1 KB | ||
| Others |  emd_33499_half_map_1.map.gz emd_33499_half_map_1.map.gz emd_33499_half_map_2.map.gz emd_33499_half_map_2.map.gz | 227.8 MB 227.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33499 http://ftp.pdbj.org/pub/emdb/structures/EMD-33499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33499 | HTTPS FTP |
-Related structure data
| Related structure data |  7xxaMC  7xxgC  7xxjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33499.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33499.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33499_msk_1.map emd_33499_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33499_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33499_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Echovirus E18
| Entire | Name:  Echovirus E18 Echovirus E18 |
|---|---|
| Components |
|
-Supramolecule #1: Echovirus E18
| Supramolecule | Name: Echovirus E18 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
-Supramolecule #3: FcRn
| Supramolecule | Name: FcRn / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Supramolecule #2: Homo sapiens
| Supramolecule | Name: Homo sapiens / type: virus / ID: 2 / Parent: 1 / Macromolecule list: #1, #3-#5 / NCBI-ID: 9606 / Sci species name: Homo sapiens / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 35.113328 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GDNQDRTVAN TQPSGPSNSK EIPALTAVET GHTSQVDPSD TLQTRHVVNF HSRSESTVEN FMGRAACVFM DQYKLNGEET STDNFAVWT INVREMAQLR RKCELFTYMR FDIEMTMVIT SCQDQGTQLE QDMPVLTHQI MYVPPGGPIP AKVDSYEWQT S TNPSVFWT ...String: GDNQDRTVAN TQPSGPSNSK EIPALTAVET GHTSQVDPSD TLQTRHVVNF HSRSESTVEN FMGRAACVFM DQYKLNGEET STDNFAVWT INVREMAQLR RKCELFTYMR FDIEMTMVIT SCQDQGTQLE QDMPVLTHQI MYVPPGGPIP AKVDSYEWQT S TNPSVFWT EGNAPARMSI PFISVGNAYS LFYDGWSHFT QDGTYGYTTL NAMGKLFVRH VNKSSPHQIT STIRVYFKPK HI KAWVPRP PRLCPYINKG DVNFVVTEVT DARKSITDTP HPEHSVLVTR GAFGQQSGAA YIGNYKVVNR HLATH |
-Macromolecule #2: IgG receptor FcRn large subunit p51
| Macromolecule | Name: IgG receptor FcRn large subunit p51 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.294971 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LSLLYHLTAV SSPAPGTPAF WVSGWLGPQQ YLSYNSLRGE AEPCGAWVWE NQVSWYWEKE TTDLRIKEKL FLEAFKALGG KGPYTLQGL LGCELGPDNT SVPTAKFALN GEEFMNFDLK QGTWGGDWPE ALAISQRWQQ QDKAANKELT FLLFSCPHRL R EHLERGRG ...String: LSLLYHLTAV SSPAPGTPAF WVSGWLGPQQ YLSYNSLRGE AEPCGAWVWE NQVSWYWEKE TTDLRIKEKL FLEAFKALGG KGPYTLQGL LGCELGPDNT SVPTAKFALN GEEFMNFDLK QGTWGGDWPE ALAISQRWQQ QDKAANKELT FLLFSCPHRL R EHLERGRG NLEWKEPPSM RLKARPSSPG FSVLTCSAFS FYPPELQLRF LRNGLAAGTG QGDFGPNSDG SFHASSSLTV KS GDEHHYC CIVQHAGLAQ PLRVEL UniProtKB: IgG receptor FcRn large subunit p51 |
-Macromolecule #3: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 28.805348 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SPSAEECGYS DRVRSMTLGN STITTQESAN VVVGYGEWPS YLSDKEATAE DQPTQPDVAT CRFYTLESVQ WEKSSPGWWW KFPEALKNM GLFGQNMHYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCADTST TFPATELTTE EEPHVFTSDS I TGKKVQAA ...String: SPSAEECGYS DRVRSMTLGN STITTQESAN VVVGYGEWPS YLSDKEATAE DQPTQPDVAT CRFYTLESVQ WEKSSPGWWW KFPEALKNM GLFGQNMHYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGCADTST TFPATELTTE EEPHVFTSDS I TGKKVQAA VCNAGMGVGV GNLTIFPHQW INLRTNNSAT IVMPYINSVP MDNMFRHYNF TLMIIPFAPL NFNEGATAYV PV TVTIAPM YAEYNGLRLA STQ |
-Macromolecule #4: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 26.134842 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GVPVLNTPGS TQFLTSDDFQ SPSAMPQFDE TPEMHIPGEV RNLMEMAEVD SVVPVNNITG KTKSMEAYQI AVGTGNTDKT KPIFSFQMD PGYSSVLKRT LLGEMLNYYA HWSGSVKLTF LFCGSAMATG KLLISYSPPG ASVPSSRKDA MLGTHIIWDI G LQSSCVLC ...String: GVPVLNTPGS TQFLTSDDFQ SPSAMPQFDE TPEMHIPGEV RNLMEMAEVD SVVPVNNITG KTKSMEAYQI AVGTGNTDKT KPIFSFQMD PGYSSVLKRT LLGEMLNYYA HWSGSVKLTF LFCGSAMATG KLLISYSPPG ASVPSSRKDA MLGTHIIWDI G LQSSCVLC VPWISQSHYR MVQQDPYTSA GYITCWYQTN IVVPPGAPTS CDVLCFASAC NDFSVRLLRD TPFMAQPGKL Q |
-Macromolecule #5: VP4
| Macromolecule | Name: VP4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Echovirus E18 Echovirus E18 |
| Molecular weight | Theoretical: 7.502346 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAQVSTQKT GAHETSLNAK GNSIIHYTNI NFYKDAASSA SNRQELQQDP GKFTDPVKDL MVKTLPALN |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot for 3 seconds and wait for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Alignment procedure | Coma free - Residual tilt: 100.0 mrad |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2028 / Average exposure time: 0.088 sec. / Average electron dose: 1.025 e/Å2 / Details: Images were collected in super-resolution mode. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||
| Output model |  PDB-7xxa: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)