[English] 日本語
 Yorodumi
Yorodumi- EMDB-33226: Cryo-EM structure of E.coli retron-Ec86 in complex with its effec... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Reverse transcriptase / RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed DNA polymerase / RNA-directed DNA polymerase activity / defense response to virus / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.51 Å | |||||||||
 Authors Authors | Wang YJ / Guan ZY / Zou TT | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism. Authors: Yanjing Wang / Zeyuan Guan / Chen Wang / Yangfan Nie / Yibei Chen / Zhaoyang Qian / Yongqing Cui / Han Xu / Qiang Wang / Fen Zhao / Delin Zhang / Pan Tao / Ming Sun / Ping Yin / Shuangxia ...Authors: Yanjing Wang / Zeyuan Guan / Chen Wang / Yangfan Nie / Yibei Chen / Zhaoyang Qian / Yongqing Cui / Han Xu / Qiang Wang / Fen Zhao / Delin Zhang / Pan Tao / Ming Sun / Ping Yin / Shuangxia Jin / Shan Wu / Tingting Zou /  Abstract: First discovered in the 1980s, retrons are bacterial genetic elements consisting of a reverse transcriptase and a non-coding RNA (ncRNA). Retrons mediate antiphage defence in bacteria but their ...First discovered in the 1980s, retrons are bacterial genetic elements consisting of a reverse transcriptase and a non-coding RNA (ncRNA). Retrons mediate antiphage defence in bacteria but their structure and defence mechanisms are unknown. Here, we investigate the Escherichia coli Ec86 retron and use cryo-electron microscopy to determine the structures of the Ec86 (3.1 Å) and cognate effector-bound Ec86 (2.5 Å) complexes. The Ec86 reverse transcriptase exhibits a characteristic right-hand-like fold consisting of finger, palm and thumb subdomains. Ec86 reverse transcriptase reverse-transcribes part of the ncRNA into satellite, multicopy single-stranded DNA (msDNA, a DNA-RNA hybrid) that we show wraps around the reverse transcriptase electropositive surface. In msDNA, both inverted repeats are present and the 3' sides of the DNA/RNA chains are close to the reverse transcriptase active site. The Ec86 effector adopts a two-lobe fold and directly binds reverse transcriptase and msDNA. These findings offer insights into the structure-function relationship of the retron-effector unit and provide a structural basis for the optimization of retron-based genome editing systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33226.map.gz emd_33226.map.gz | 111.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33226-v30.xml emd-33226-v30.xml emd-33226.xml emd-33226.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33226.png emd_33226.png | 105.1 KB | ||
| Filedesc metadata |  emd-33226.cif.gz emd-33226.cif.gz | 6.8 KB | ||
| Others |  emd_33226_half_map_1.map.gz emd_33226_half_map_1.map.gz emd_33226_half_map_2.map.gz emd_33226_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33226 http://ftp.pdbj.org/pub/emdb/structures/EMD-33226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33226 | HTTPS FTP |
-Related structure data
| Related structure data |  7xjgMC  7v9uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33226.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33226.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
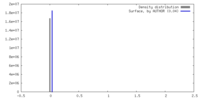
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33226_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
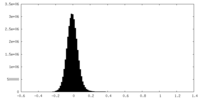
| Density Histograms |
-Half map: #1
| File | emd_33226_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
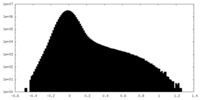
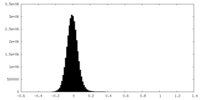
| Density Histograms |
- Sample components
Sample components
-Entire : retron-Ec86 (RT-msDNA-RNA)
| Entire | Name: retron-Ec86 (RT-msDNA-RNA) |
|---|---|
| Components |
|
-Supramolecule #1: retron-Ec86 (RT-msDNA-RNA)
| Supramolecule | Name: retron-Ec86 (RT-msDNA-RNA) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RNA-directed DNA polymerase from retron EC86
| Macromolecule | Name: RNA-directed DNA polymerase from retron EC86 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.83207 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSAEYLNTF RLRNLGLPVM NNLHDMSKAT RISVETLRLL IYTADFRYRI YTVEKKGPEK RMRTIYQPSR ELKALQGWVL RNILDKLSS SPFSIGFEKH QSILNNATPH IGANFILNID LEDFFPSLTA NKVFGVFHSL GYNRLISSVL TKICCYKNLL P QGAPSSPK ...String: MKSAEYLNTF RLRNLGLPVM NNLHDMSKAT RISVETLRLL IYTADFRYRI YTVEKKGPEK RMRTIYQPSR ELKALQGWVL RNILDKLSS SPFSIGFEKH QSILNNATPH IGANFILNID LEDFFPSLTA NKVFGVFHSL GYNRLISSVL TKICCYKNLL P QGAPSSPK LANLICSKLD YRIQGYAGSR GLIYTRYADD LTLSAQSMKK VVKARDFLFS IIPSEGLVIN SKKTCISGPR SQ RKVTGLV ISQEKVGIGR EKYKEIRAKI HHIFCGKSSE IEHVRGWLSF ILSVDSKSHR RLITYISKLE KKYGKNPLNK AKT LEHHHH HHHH UniProtKB: Retron Ec86 reverse transcriptase |
-Macromolecule #2: retron St85 family effector protein
| Macromolecule | Name: retron St85 family effector protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.538242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNKKFTDEQQ QQLIGHLTKK GFYRGANIKI TIFLCGGDVA NHQSWRHQLS QFLAKFSDVD IFYPEDLFDD LLAGQGQHSL LSLENILAE AVDVIILFPE SPGSFTELGA FSNNENLRRK LICIQDAKFK SKRSFINYGP VRLLRKFNSK SVLRCSSNEL K EMCDSSID ...String: MNKKFTDEQQ QQLIGHLTKK GFYRGANIKI TIFLCGGDVA NHQSWRHQLS QFLAKFSDVD IFYPEDLFDD LLAGQGQHSL LSLENILAE AVDVIILFPE SPGSFTELGA FSNNENLRRK LICIQDAKFK SKRSFINYGP VRLLRKFNSK SVLRCSSNEL K EMCDSSID VARKLRLYKK LMASIKKVRK ENKVSKDIGN ILYAERFLLP CIYLLDSVNY RTLCELAFKA IKQDDVLSKI IV RSVVSRL INERKILQMT DGYQVTALGA SYVRSVFDRK TLDRLRLEIM NFENRRKSTF NYDKIPYAHP UniProtKB: UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase |
-Macromolecule #3: DNA (105-MER)
| Macromolecule | Name: DNA (105-MER) / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.468803 KDa |
| Sequence | String: (DG)(DA)(DA)(DA)(DG)(DT)(DT)(DG)(DC)(DG) (DC)(DA)(DC)(DC)(DC)(DT)(DT)(DA)(DC)(DG) (DT)(DC)(DA)(DG)(DA)(DA)(DA)(DA)(DA) (DA)(DC)(DG)(DG)(DG)(DT)(DT)(DT)(DC)(DC) (DT) (DG)(DG)(DT)(DT)(DG)(DG) ...String: (DG)(DA)(DA)(DA)(DG)(DT)(DT)(DG)(DC)(DG) (DC)(DA)(DC)(DC)(DC)(DT)(DT)(DA)(DC)(DG) (DT)(DC)(DA)(DG)(DA)(DA)(DA)(DA)(DA) (DA)(DC)(DG)(DG)(DG)(DT)(DT)(DT)(DC)(DC) (DT) (DG)(DG)(DT)(DT)(DG)(DG)(DC)(DT) (DC)(DG)(DG)(DA)(DG)(DA)(DG)(DC)(DA)(DT) (DC)(DA) (DG)(DG)(DC)(DG)(DA)(DT)(DG) (DC)(DT)(DC)(DT)(DC)(DC)(DG)(DT)(DT)(DC) (DC)(DA)(DA) (DC)(DA)(DA)(DG)(DG)(DA) (DA)(DA)(DA)(DC)(DA)(DG)(DA)(DC)(DA)(DG) (DT)(DA)(DA)(DC) (DT)(DC)(DA)(DG)(DA) GENBANK: GENBANK: M24408.1 |
-Macromolecule #4: RNA (81-MER)
| Macromolecule | Name: RNA (81-MER) / type: rna / ID: 4 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.870203 KDa |
| Sequence | String: UGCGCACCCU UAGCGAGAGG UUUAUCAUUA AGGUCAACCU CUGGAUGUUG UUUCGGCAUC CUGCAUUGAA UCUGAGUUAC U GENBANK: GENBANK: M24408.1 |
-Macromolecule #5: RNA (14-MER)
| Macromolecule | Name: RNA (14-MER) / type: rna / ID: 5 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.541771 KDa |
| Sequence | String: CGUAAGGGUG CGCA |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)