+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
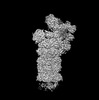
| タイトル | Structure of USP14-bound human 26S proteasome in state EA1_UBL | |||||||||
 マップデータ マップデータ | Complete map of the human proteasome holoenzyme in state EA1_UBL, with the RP/CP low-pass filtered to 3.3/2.8 angstrom and sharpened by a B-factor of -30/0 | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Proteasome / AAA-ATPase / Deubiquitinase / USP14 / HYDROLASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of ERAD pathway / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / positive regulation of inclusion body assembly / regulation of chemotaxis / protein K48-linked deubiquitination / deubiquitinase activity ...negative regulation of ERAD pathway / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / positive regulation of inclusion body assembly / regulation of chemotaxis / protein K48-linked deubiquitination / deubiquitinase activity / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; オメガペプチターゼ / proteasome accessory complex / integrator complex / purine ribonucleoside triphosphate binding / meiosis I / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process / proteasome-activating activity / proteasome regulatory particle, lid subcomplex / proteasome regulatory particle, base subcomplex / metal-dependent deubiquitinase activity / negative regulation of programmed cell death / protein K63-linked deubiquitination / Regulation of ornithine decarboxylase (ODC) / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / proteasome core complex / Resolution of D-loop Structures through Holliday Junction Intermediates / Somitogenesis / endopeptidase inhibitor activity / K63-linked deubiquitinase activity / Impaired BRCA2 binding to RAD51 / proteasome binding / transcription factor binding / regulation of protein catabolic process / myofibril / proteasome storage granule / Presynaptic phase of homologous DNA pairing and strand exchange / general transcription initiation factor binding / blastocyst development / polyubiquitin modification-dependent protein binding / negative regulation of ubiquitin-dependent protein catabolic process / protein deubiquitination / immune system process / endopeptidase activator activity / NF-kappaB binding / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome assembly / threonine-type endopeptidase activity / proteasome core complex, alpha-subunit complex / mRNA export from nucleus / presynaptic cytosol / enzyme regulator activity / inclusion body / regulation of proteasomal protein catabolic process / proteasome complex / TBP-class protein binding / proteolysis involved in protein catabolic process / sarcomere / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Asymmetric localization of PCP proteins / Regulation of NF-kappa B signaling / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / stem cell differentiation / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA / Vpu mediated degradation of CD4 / Assembly of the pre-replicative complex / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Degradation of DVL / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Dectin-1 mediated noncanonical NF-kB signaling / lipopolysaccharide binding / negative regulation of inflammatory response to antigenic stimulus / Degradation of AXIN / Hh mutants are degraded by ERAD / Activation of NF-kappaB in B cells / P-body / Degradation of GLI1 by the proteasome / Hedgehog ligand biogenesis / G2/M Checkpoints / Defective CFTR causes cystic fibrosis / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Autodegradation of the E3 ubiquitin ligase COP1 / Negative regulation of NOTCH4 signaling / Regulation of RUNX3 expression and activity / Vif-mediated degradation of APOBEC3G / Hedgehog 'on' state 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.0 Å | |||||||||
 データ登録者 データ登録者 | Zhang S / Zou S / Yin D / Wu Z / Mao Y | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2022 ジャーナル: Nature / 年: 2022タイトル: USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. 著者: Shuwen Zhang / Shitao Zou / Deyao Yin / Lihong Zhao / Daniel Finley / Zhaolong Wu / Youdong Mao /   要旨: Proteasomal degradation of ubiquitylated proteins is tightly regulated at multiple levels. A primary regulatory checkpoint is the removal of ubiquitin chains from substrates by the deubiquitylating ...Proteasomal degradation of ubiquitylated proteins is tightly regulated at multiple levels. A primary regulatory checkpoint is the removal of ubiquitin chains from substrates by the deubiquitylating enzyme ubiquitin-specific protease 14 (USP14), which reversibly binds the proteasome and confers the ability to edit and reject substrates. How USP14 is activated and regulates proteasome function remain unknown. Here we present high-resolution cryo-electron microscopy structures of human USP14 in complex with the 26S proteasome in 13 distinct conformational states captured during degradation of polyubiquitylated proteins. Time-resolved cryo-electron microscopy analysis of the conformational continuum revealed two parallel pathways of proteasome state transitions induced by USP14, and captured transient conversion of substrate-engaged intermediates into substrate-inhibited intermediates. On the substrate-engaged pathway, ubiquitin-dependent activation of USP14 allosterically reprograms the conformational landscape of the AAA-ATPase motor and stimulates opening of the core particle gate, enabling observation of a near-complete cycle of asymmetric ATP hydrolysis around the ATPase ring during processive substrate unfolding. Dynamic USP14-ATPase interactions decouple the ATPase activity from RPN11-catalysed deubiquitylation and kinetically introduce three regulatory checkpoints on the proteasome, at the steps of ubiquitin recognition, substrate translocation initiation and ubiquitin chain recycling. These findings provide insights into the complete functional cycle of the USP14-regulated proteasome and establish mechanistic foundations for the discovery of USP14-targeted therapies. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_32272.map.gz emd_32272.map.gz | 887.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-32272-v30.xml emd-32272-v30.xml emd-32272.xml emd-32272.xml | 64.8 KB 64.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_32272.png emd_32272.png | 76.4 KB | ||
| Filedesc metadata |  emd-32272.cif.gz emd-32272.cif.gz | 14.8 KB | ||
| その他 |  emd_32272_additional_1.map.gz emd_32272_additional_1.map.gz emd_32272_additional_2.map.gz emd_32272_additional_2.map.gz emd_32272_additional_3.map.gz emd_32272_additional_3.map.gz emd_32272_additional_4.map.gz emd_32272_additional_4.map.gz | 868.6 MB 893.3 MB 870.7 MB 892.2 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32272 http://ftp.pdbj.org/pub/emdb/structures/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32272 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_32272_validation.pdf.gz emd_32272_validation.pdf.gz | 579 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_32272_full_validation.pdf.gz emd_32272_full_validation.pdf.gz | 578.5 KB | 表示 | |
| XML形式データ |  emd_32272_validation.xml.gz emd_32272_validation.xml.gz | 8.5 KB | 表示 | |
| CIF形式データ |  emd_32272_validation.cif.gz emd_32272_validation.cif.gz | 9.8 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32272 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32272 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7w37MC  7w38C  7w39C  7w3aC  7w3bC  7w3cC  7w3fC  7w3gC  7w3hC  7w3iC  7w3jC  7w3kC  7w3mC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_32272.map.gz / 形式: CCP4 / 大きさ: 1000 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_32272.map.gz / 形式: CCP4 / 大きさ: 1000 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Complete map of the human proteasome holoenzyme in state EA1_UBL, with the RP/CP low-pass filtered to 3.3/2.8 angstrom and sharpened by a B-factor of -30/0 | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.685 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: Unfiltered, unsharpened raw map of the human proteasome...
| ファイル | emd_32272_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unfiltered, unsharpened raw map of the human proteasome in state EA1_UBL after RP-masked refinement | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
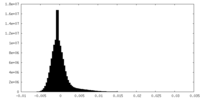
| 密度ヒストグラム |
-追加マップ: Map of the human proteasome in state EA1 UBL...
| ファイル | emd_32272_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Map of the human proteasome in state EA1_UBL after RP-masked refinement, low-pass filtered to 3.3 angstrom and sharpened by a B-factor of -30 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
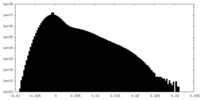
| 密度ヒストグラム |
-追加マップ: Unfiltered, unsharpened raw map of the human proteasome...
| ファイル | emd_32272_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unfiltered, unsharpened raw map of the human proteasome in state EA1_UBL after CP-masked refinement | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
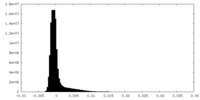
| 密度ヒストグラム |
-追加マップ: Map of the human proteasome in state EA1 UBL...
| ファイル | emd_32272_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Map of the human proteasome in state EA1_UBL after CP-masked refinement, low-pass filtered to 3.3 angstrom with no sharpening B-factor applied | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
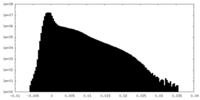
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : 26S proteasome
+超分子 #1: 26S proteasome
+分子 #1: 26S protease regulatory subunit 7
+分子 #2: 26S protease regulatory subunit 4
+分子 #3: Isoform 2 of 26S proteasome regulatory subunit 8
+分子 #4: 26S protease regulatory subunit 6B
+分子 #5: 26S proteasome regulatory subunit 10B
+分子 #6: 26S protease regulatory subunit 6A
+分子 #7: Proteasome subunit alpha type-6
+分子 #8: Proteasome subunit alpha type-2
+分子 #9: Proteasome subunit alpha type-4
+分子 #10: Proteasome subunit alpha type-7
+分子 #11: Proteasome subunit alpha type-5
+分子 #12: Isoform Long of Proteasome subunit alpha type-1
+分子 #13: Proteasome subunit alpha type-3
+分子 #14: Proteasome subunit beta type-6
+分子 #15: Proteasome subunit beta type-7
+分子 #16: Proteasome subunit beta type-3
+分子 #17: Proteasome subunit beta type-2
+分子 #18: Proteasome subunit beta type-5
+分子 #19: Proteasome subunit beta type-1
+分子 #20: Proteasome subunit beta type-4
+分子 #21: 26S proteasome non-ATPase regulatory subunit 1
+分子 #22: 26S proteasome non-ATPase regulatory subunit 3
+分子 #23: 26S proteasome non-ATPase regulatory subunit 12
+分子 #24: 26S proteasome non-ATPase regulatory subunit 11
+分子 #25: 26S proteasome non-ATPase regulatory subunit 6
+分子 #26: 26S proteasome non-ATPase regulatory subunit 7
+分子 #27: 26S proteasome non-ATPase regulatory subunit 13
+分子 #28: 26S proteasome non-ATPase regulatory subunit 4
+分子 #29: 26S proteasome non-ATPase regulatory subunit 14
+分子 #30: 26S proteasome non-ATPase regulatory subunit 8
+分子 #31: 26S proteasome non-ATPase regulatory subunit 2
+分子 #32: Ubiquitin carboxyl-terminal hydrolase 14
+分子 #33: 26S proteasome complex subunit DSS1
+分子 #34: ADENOSINE-5'-TRIPHOSPHATE
+分子 #35: MAGNESIUM ION
+分子 #36: ADENOSINE-5'-DIPHOSPHATE
+分子 #37: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 5.0 µm / 最小 デフォーカス(公称値): 0.4 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: EMDB MAP EMDB ID: |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.0 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 367235 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)