+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of USP14-bound human 26S proteasome in state EA1_UBL | |||||||||
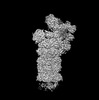
 Map data Map data | Complete map of the human proteasome holoenzyme in state EA1_UBL, with the RP/CP low-pass filtered to 3.3/2.8 angstrom and sharpened by a B-factor of -30/0 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Proteasome / AAA-ATPase / Deubiquitinase / USP14 / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of ERAD pathway / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / deubiquitinase activity / regulation of chemotaxis / protein K48-linked deubiquitination ...negative regulation of ERAD pathway / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / thyrotropin-releasing hormone receptor binding / nuclear proteasome complex / host-mediated perturbation of viral transcription / positive regulation of inclusion body assembly / deubiquitinase activity / regulation of chemotaxis / protein K48-linked deubiquitination / Hydrolases; Acting on peptide bonds (peptidases); Omega peptidases / integrator complex / proteasome accessory complex / meiosis I / purine ribonucleoside triphosphate binding / proteasome regulatory particle / cytosolic proteasome complex / positive regulation of proteasomal protein catabolic process / proteasome-activating activity / proteasome regulatory particle, lid subcomplex / proteasome regulatory particle, base subcomplex / protein K63-linked deubiquitination / metal-dependent deubiquitinase activity / negative regulation of programmed cell death / Regulation of ornithine decarboxylase (ODC) / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / proteasome core complex / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Somitogenesis / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / endopeptidase inhibitor activity / K63-linked deubiquitinase activity / Resolution of D-loop Structures through Holliday Junction Intermediates / proteasome binding / Impaired BRCA2 binding to RAD51 / transcription factor binding / regulation of protein catabolic process / myofibril / proteasome storage granule / Presynaptic phase of homologous DNA pairing and strand exchange / general transcription initiation factor binding / polyubiquitin modification-dependent protein binding / protein deubiquitination / blastocyst development / negative regulation of ubiquitin-dependent protein catabolic process / immune system process / NF-kappaB binding / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / endopeptidase activator activity / proteasome assembly / threonine-type endopeptidase activity / mRNA export from nucleus / proteasome core complex, alpha-subunit complex / presynaptic cytosol / enzyme regulator activity / regulation of proteasomal protein catabolic process / inclusion body / proteasome complex / : / TBP-class protein binding / sarcomere / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / Regulation of NF-kappa B signaling / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / stem cell differentiation / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Ubiquitin-dependent degradation of Cyclin D / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA / Vpu mediated degradation of CD4 / Assembly of the pre-replicative complex / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / negative regulation of inflammatory response to antigenic stimulus / Degradation of DVL / Dectin-1 mediated noncanonical NF-kB signaling / P-body / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Degradation of AXIN / lipopolysaccharide binding / Hh mutants are degraded by ERAD / Activation of NF-kappaB in B cells / Degradation of GLI1 by the proteasome / G2/M Checkpoints / Hedgehog ligand biogenesis / Autodegradation of the E3 ubiquitin ligase COP1 / Defective CFTR causes cystic fibrosis / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Regulation of RUNX3 expression and activity / Negative regulation of NOTCH4 signaling / Hedgehog 'on' state / Vif-mediated degradation of APOBEC3G Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhang S / Zou S / Yin D / Wu Z / Mao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Authors: Shuwen Zhang / Shitao Zou / Deyao Yin / Lihong Zhao / Daniel Finley / Zhaolong Wu / Youdong Mao /   Abstract: Proteasomal degradation of ubiquitylated proteins is tightly regulated at multiple levels. A primary regulatory checkpoint is the removal of ubiquitin chains from substrates by the deubiquitylating ...Proteasomal degradation of ubiquitylated proteins is tightly regulated at multiple levels. A primary regulatory checkpoint is the removal of ubiquitin chains from substrates by the deubiquitylating enzyme ubiquitin-specific protease 14 (USP14), which reversibly binds the proteasome and confers the ability to edit and reject substrates. How USP14 is activated and regulates proteasome function remain unknown. Here we present high-resolution cryo-electron microscopy structures of human USP14 in complex with the 26S proteasome in 13 distinct conformational states captured during degradation of polyubiquitylated proteins. Time-resolved cryo-electron microscopy analysis of the conformational continuum revealed two parallel pathways of proteasome state transitions induced by USP14, and captured transient conversion of substrate-engaged intermediates into substrate-inhibited intermediates. On the substrate-engaged pathway, ubiquitin-dependent activation of USP14 allosterically reprograms the conformational landscape of the AAA-ATPase motor and stimulates opening of the core particle gate, enabling observation of a near-complete cycle of asymmetric ATP hydrolysis around the ATPase ring during processive substrate unfolding. Dynamic USP14-ATPase interactions decouple the ATPase activity from RPN11-catalysed deubiquitylation and kinetically introduce three regulatory checkpoints on the proteasome, at the steps of ubiquitin recognition, substrate translocation initiation and ubiquitin chain recycling. These findings provide insights into the complete functional cycle of the USP14-regulated proteasome and establish mechanistic foundations for the discovery of USP14-targeted therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32272.map.gz emd_32272.map.gz | 887.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32272-v30.xml emd-32272-v30.xml emd-32272.xml emd-32272.xml | 64.8 KB 64.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32272.png emd_32272.png | 76.4 KB | ||
| Filedesc metadata |  emd-32272.cif.gz emd-32272.cif.gz | 14.8 KB | ||
| Others |  emd_32272_additional_1.map.gz emd_32272_additional_1.map.gz emd_32272_additional_2.map.gz emd_32272_additional_2.map.gz emd_32272_additional_3.map.gz emd_32272_additional_3.map.gz emd_32272_additional_4.map.gz emd_32272_additional_4.map.gz | 868.6 MB 893.3 MB 870.7 MB 892.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32272 http://ftp.pdbj.org/pub/emdb/structures/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32272 | HTTPS FTP |
-Related structure data
| Related structure data |  7w37MC  7w38C  7w39C  7w3aC  7w3bC  7w3cC  7w3fC  7w3gC  7w3hC  7w3iC  7w3jC  7w3kC  7w3mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32272.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32272.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complete map of the human proteasome holoenzyme in state EA1_UBL, with the RP/CP low-pass filtered to 3.3/2.8 angstrom and sharpened by a B-factor of -30/0 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.685 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
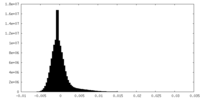
-Additional map: Unfiltered, unsharpened raw map of the human proteasome...
| File | emd_32272_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered, unsharpened raw map of the human proteasome in state EA1_UBL after RP-masked refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
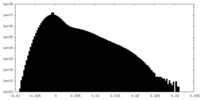
-Additional map: Map of the human proteasome in state EA1 UBL...
| File | emd_32272_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the human proteasome in state EA1_UBL after RP-masked refinement, low-pass filtered to 3.3 angstrom and sharpened by a B-factor of -30 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
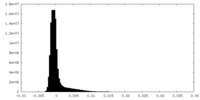
-Additional map: Unfiltered, unsharpened raw map of the human proteasome...
| File | emd_32272_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered, unsharpened raw map of the human proteasome in state EA1_UBL after CP-masked refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
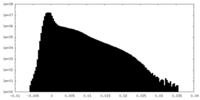
-Additional map: Map of the human proteasome in state EA1 UBL...
| File | emd_32272_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the human proteasome in state EA1_UBL after CP-masked refinement, low-pass filtered to 3.3 angstrom with no sharpening B-factor applied | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 26S proteasome
+Supramolecule #1: 26S proteasome
+Macromolecule #1: 26S protease regulatory subunit 7
+Macromolecule #2: 26S protease regulatory subunit 4
+Macromolecule #3: Isoform 2 of 26S proteasome regulatory subunit 8
+Macromolecule #4: 26S protease regulatory subunit 6B
+Macromolecule #5: 26S proteasome regulatory subunit 10B
+Macromolecule #6: 26S protease regulatory subunit 6A
+Macromolecule #7: Proteasome subunit alpha type-6
+Macromolecule #8: Proteasome subunit alpha type-2
+Macromolecule #9: Proteasome subunit alpha type-4
+Macromolecule #10: Proteasome subunit alpha type-7
+Macromolecule #11: Proteasome subunit alpha type-5
+Macromolecule #12: Isoform Long of Proteasome subunit alpha type-1
+Macromolecule #13: Proteasome subunit alpha type-3
+Macromolecule #14: Proteasome subunit beta type-6
+Macromolecule #15: Proteasome subunit beta type-7
+Macromolecule #16: Proteasome subunit beta type-3
+Macromolecule #17: Proteasome subunit beta type-2
+Macromolecule #18: Proteasome subunit beta type-5
+Macromolecule #19: Proteasome subunit beta type-1
+Macromolecule #20: Proteasome subunit beta type-4
+Macromolecule #21: 26S proteasome non-ATPase regulatory subunit 1
+Macromolecule #22: 26S proteasome non-ATPase regulatory subunit 3
+Macromolecule #23: 26S proteasome non-ATPase regulatory subunit 12
+Macromolecule #24: 26S proteasome non-ATPase regulatory subunit 11
+Macromolecule #25: 26S proteasome non-ATPase regulatory subunit 6
+Macromolecule #26: 26S proteasome non-ATPase regulatory subunit 7
+Macromolecule #27: 26S proteasome non-ATPase regulatory subunit 13
+Macromolecule #28: 26S proteasome non-ATPase regulatory subunit 4
+Macromolecule #29: 26S proteasome non-ATPase regulatory subunit 14
+Macromolecule #30: 26S proteasome non-ATPase regulatory subunit 8
+Macromolecule #31: 26S proteasome non-ATPase regulatory subunit 2
+Macromolecule #32: Ubiquitin carboxyl-terminal hydrolase 14
+Macromolecule #33: 26S proteasome complex subunit DSS1
+Macromolecule #34: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #35: MAGNESIUM ION
+Macromolecule #36: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #37: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 367235 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)